Codementor: Why you should contribute to Open Source
Codementor: Supporting Multiple Languages In Django — Part 1
John Cook: Solving Van der Pol equation with ivp_solve
Van der Pol’s differential equation is
The equation describes a system with nonlinear damping, the degree of damping given by μ. If μ = 0 the system is linear and undamped, but for positive μ the system is nonlinear and damped. We will plot the phase portrait for the solution to Van der Pol’s equation in Python using SciPy’s new ODE solver ivp_solve.
The function ivp_solve does not solve second-order systems of equations directly. It solves systems of first-order equations, but a second-order differential equation can be recast as a pair of first-order equations by introducing the first derivative as a new variable.
Since y is the derivative of x, the phase portrait is just the plot of (x, y).
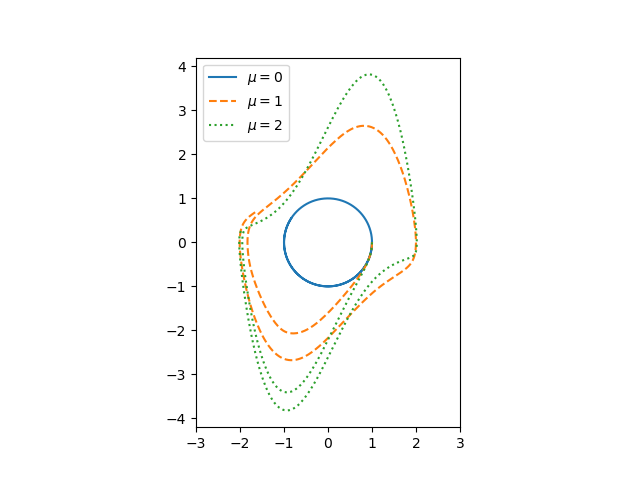
If μ = 0, we have a simple harmonic oscillator and the phase portrait is simply a circle. For larger values of μ the solutions enter limiting cycles, but the cycles are more complicated than just circles.
Here’s the Python code that made the plot.
from scipy import linspace
from scipy.integrate import solve_ivp
import matplotlib.pyplot as plt
def vdp(t, z):
x, y = z
return [y, mu*(1 - x**2)*y - x]
a, b = 0, 10
mus = [0, 1, 2]
styles = ["-", "--", ":"]
t = linspace(a, b, 1000)
for mu, style in zip(mus, styles):
sol = solve_ivp(vdp, [a, b], [1, 0], t_eval=t)
plt.plot(sol.y[0], sol.y[1], style)
# make a little extra horizontal room for legend
plt.xlim([-3,3])
plt.legend([f"$\mu={m}$" for m in mus])
plt.axes().set_aspect(1)
Related posts
Mike Driscoll: PyDev of the Week: Kelly Schuster-Paredes
This week we welcome Kelly (@KellyPared) Schuster-Paredes. Kelly is the co-host of the popular Python podcast, Teaching Python. Kelly specializes in curriculum design and development. She blogs a bit over on her website which you should check out if you have the time.
For now though, let’s take a few moments to get to know Kelly better!
Can you tell us a little about yourself (hobbies, education, etc):
I am a Middle School Computer Science Teacher and a Technology Integration Specialist. I have been teaching for 23 years and have taught in the US, UK and in Peru. I have a Masters in Curriculum, Instruction and Technology, which means I know a lot about how to teach and invent cool lessons. Besides working and co-hosting Teaching Python, I spend most of my time with my two boys outside playing sports in the south Florida sun.
Why did you start using Python?
I started using Python almost two years ago. My boss told me in April 2018 that I was going to be teaching Python to middle school kids in August of that year. And I said, “Over my dead body I am learning Python, why not Javascript?” Needless to say, I didn’t win the battle, thankfully! Seriously though, my boss knew I loved challenges and will take on anything presented to me and then make it awesome. I believe wholeheartedly, she made the right choice.
What other programming languages do you know and which is your favorite?
I “know” HTML and what most educators call Block Code, and that is the extent of my coding language. I can write a few lines of Javascript and a few lines of SWIFT but Python is my only language that I can communicate in somewhat fluently.
What projects are you working on now?
I do not have any major projects formulated currently. Do to my role and how I teach in computer science, I am constantly having to know a little about everything. We allow our students to follow their passions, so at any one moment in time I may be helping a student with circuitpython, micropython, pygame, matplotlib, tensorflow, raspberrypi, turtle, pandas, and even libraries like requests, and translate.
I am dreaming of developing a program that can act as a “dashboard” to analyze all points of data on a student, from grades, attendance, homelife, standardized test scores, academic behavior, you name it. However, this will take quite a bit more time than I have at the moment, so I currently just have the start of the ‘database’ aspect. I hope to find time one day to put in the full effort to make it happen. I am also in the process of writing a book, called Code Like a Native: Becoming a Polyglot, but this too is a very slow process and I hope to get this done in the near future.
Which Python libraries are your favorite (core or 3rd party)?
I am a former pre-med student and love everything about science and math. I love graphs and seeing things visually. I think my favorite libraries are the ones that deal with Data Science. I especially like MatPlotLib because of its readability and simplicity to make amazing graphs.
I also have to say, I have a love hate relationship with CIrcuitPython. I really like fashion design and the possibilities of designing some really cool tech fashionware keeps me hooked. I watch some of the pythonistas on Twitter with their cool color changing fairy wings and LED patterned dresses and think, “Oooh one day, that is going to be one of my students creating that!”
How did your podcast, Teaching Python, come about?
Teaching Python came about because of the amazing teaching partner and friend that I have, Sean Tibor. Sean and I started working together almost two years ago. He has a brilliant mind and a big caring heart. I was his mentor as a new teacher and he quickly became my mentor in learning code. We started having these very deep, theoretical and critical conversations about teaching and learning python and one day we said, we need to record these!
There is a strong passion for teaching and learning to code that we felt more people should hear about what we do. We teach 370 kids a year how to code and we take 10 year old students who only know Scratch and get them making cool Python projects in less than 9 weeks. We thought, “Wow, this can be shared and more people can learn to code!”
Sean is also into digital marketing and he said, “Let’s make a podcast!”. I laughed, but we did and 36 episodes and 96,000+ downloads later, here we are.
We originally thought that our listeners would be only computer science or STEM teachers but we are realizing that we are also reaching adult, college students and other newbie coders and this is super cool for us. We have interviewed a variety of people, from former teachers, new authors, college professors and even agriculturalist! I am hoping that I can help other teachers like me make the change or take the risk and start to learn something new.
Favorite episodes?
I really like the episodes where we dive into the teaching and reflecting aspect of learning a coding language. I think episode 23 was a very open, honest and has helped me plan out and set a few goals for this year.
What have you learned as a podcaster?
We have learned so much as podcasters! From our first recording with the fuzzy background noise to where we are today, there have been a lot of bloopers. Thank goodness for editing. We record our podcasts with just an outline of topics and our conversations are live and typically unstructured. Sometimes Sean says things before I do and I am like, “oh crap, pause, I have nothing to say on that topic” and then Sean is patient and then we move on. We constantly learn from each other even during recordings.
Because of our recent new sponsors, we have now been able to purchase new microphones and recording hardware. We are also able to outsource some of our editing. This was a huge learning moment. Michael Kennedy and Brian Okken have been helpful in giving advice on what to use to record and things like that. They have been our podcast “mentors” from the beginning and supportive of our show.
Do you have any advice for others who might be interested in podcasting or blogging?
If I had to give my top five advice tips on starting a podcast they would be:
- Just start recording don’t talk about it, just do it. It does not hurt to record and put it out there, what is the worst that can happen?
- Ask for help! There are so many people that know so much more than you do at any specific moment in time. Do not feel that you have to know everything.
- Pick something that you love to talk about and that you can never exhaust all the topics. Sean and I have an endless list of topics that we seem to not be able to get through.
- Be deliberate about it all. Set a time to record, get a specific place to host, and make it happen. Be intentional and focused on what and who you are talking to everytime you record.
- Enjoy and have fun while recording, if it becomes something you ‘have to’ do and not something you ‘want to’ do, your listeners will know and your podcast will be boring.
Is there anything else you’d like to say?
Learning python has really changed my way of learning. It has opened the doors to a lot of new things in my life. As a veteran teacher, I can say that it is critical to never stop learning. I started out as a Biology teacher at 22 years of age, got into helping teachers use computers and teaching Computational skills to students at 28 and this all helped me to move to where I am today. Understanding how to code is a very important skill to have, not because I am going to be a successful programmer later in life, but because it has trained my brain to make new connections and to solve really hard problems. And that is important.
Learning Python has also introduced me to a really great community of people. I think the Python community is one of the most caring, empathetic and open communities I have met. Whether it is your first day of learning Python or you 28th year, everyone accepts you and is willing to help you solve your coding problems or answer questions.
Thanks for doing the interview, Kelly!
The post PyDev of the Week: Kelly Schuster-Paredes appeared first on The Mouse Vs. The Python.
IslandT: Create a python function which will merge, sort and remove duplicate values from lists
In this example, we will create a single python function that will accept unlimited lists of numbers and merges them into a single list that contains sorted values as well as no duplicated values. This question is from Codewars which I have further modified it to accept unlimited lists instead of two.
def merge_arrays(*kwargs):
merge_list = []
for li in kwargs:
merge_list += li
merge_list = list(dict.fromkeys(merge_list))
merge_list.sort()
return merge_list
Run the above code with the below line of statement and see the outcome by yourself.
print(merge_arrays([1,2,3,5,6,1], [1,3,5], [9,0]))
If you have a better idea, don’t forget to leave them under the comment box below!
Chris Moffitt: Creating Interactive Dashboards from Jupyter Notebooks
Introduction
I am pleased to have another guest post from Duarte O.Carmo. He wrote series of posts in July on report generation with Papermill that were very well received. In this article, he will explore how to use Voilà and Plotly Express to convert a Jupyter notebook into a standalone interactive web site. In addition, this article will show examples of collecting data through an API endpoint, performing sentiment analysis on that data and show multiple approaches to deploying the dashboard.
About Duarte
Hey everyone! My name is Duarte O.Carmo and I’m a Consultant working
at Jabra that loves working with python and data. Make sure to visit my
website if you want to find more about me 
Since this is a long article, here are the table of contents for easier navigation:
The Goal
Jupyter notebooks are one of my favorite tools to work with data, they are simple to use, fast to set up, and flexible. However, they do have their disadvantages: source control, collaboration, and reproducibility are just some of them. As I illustrated in my prior post, I tend to enjoy seeing what I can accomplish with them.
An increasing need is the sharing of our notebooks. Sure, you can export your notebooks to html, pdf, or even use something like nbviewer to share them. But what if your data changes constantly? What if every time you run your notebook, you expect to see something different? How can you go about sharing something like that?
But what if your data changes constantly? What if every time you run your notebook, you expect to see something different? How can you go about sharing something like that?
In this article, I’ll show you how to create a Jupyter Notebook that fetches live data, builds an interactive plot and then how to deploy it as a live dashboard. When you want to share the dashboard, all you need to share with someone is a link.
Let’s have some fun with the data first.
Getting live Reddit data
We will use Reddit as the source of data for our dashboard. Reddit is a tremendous source of information, and there are a million ways to get access to it. One of my favorite ways to access the data is through a small API called pushshift. The documentation is right here.
Let’s say you wanted the most recent comments mentioning the word “python”. In python, you could use requests to get a json version of the data:
importrequestsurl="https://api.pushshift.io/reddit/search/comment/?q=python"request=requests.get(url)json_response=request.json()You can add a multitude of parameters to this request, such as:
- in a certain subreddit
- after a certain day
- sorted by up votes
- many more
To make my life easier, I built a function that allows me to call this API as a function:
defget_pushshift_data(data_type,**kwargs):""" Gets data from the pushshift api. data_type can be 'comment' or 'submission' The rest of the args are interpreted as payload. Read more: https://github.com/pushshift/api"""base_url=f"https://api.pushshift.io/reddit/search/{data_type}/"payload=kwargsrequest=requests.get(base_url,params=payload)returnrequest.json()Using the
payload
parameter and
kwargs
I can then add any payload I wish as a function. For example,
get_pushshift_data(data_type="comment",# give me commentsq="python",# that mention 'python'after="48h",# in the last 48 hourssize=1000,# maximum 1000 commentssort_type="score",# sort them by scoresort="desc")# sort descendingreturns the json response. Pretty sweet right?
Analyzing the data with Plotly Express
In what subreddits does the word ‘python’ appear more?
To answer the above question, we start by getting the data with our function:
data=get_pushshift_data(data_type="comment",q="python",after="48h",size=1000,aggs="subreddit")The
aggs
keyword asks pushshift to return an aggregation into subreddits, which basically means, group the results by subreddit.
(read about it in the documentation)
Since the json response is pretty nested, we’ll need to navigate a bit inside of the dictionary.
data=data.get("aggs").get("subreddit")And we transform the list of dictionaries returned into a pandas DataFrame, and get the top 10.
df=pandas.DataFrame.from_records(data)[0:10]Here’s what our DataFrame looks like:
| doc_count | key | |
|---|---|---|
| 0 | 352 | learnpython |
| 1 | 220 | AskReddit |
| 2 | 177 | Python |
| 3 | 139 | learnprogramming |
These are the names of the subreddits where the word
python
appears most frequently in their comments 
 !
!
Let’s plot our results with the Ploty Express library. Plotly Express is great, in my opinion, if you want to:
- create figures quickly.
![🚅]()
- create figures that are a bit more interactive than matplotlib.
![👋]()
- don’t mind a bit more installation and (imo) a bit less documentation.
![😰]()
Here’s all the code you need:
importplotly.expressaspxpx.bar(df,# our dataframex="key",# x will be the 'key' column of the dataframey="doc_count",# y will be the 'doc_count' column of the dataframetitle=f"Subreddits with most activity - comments with 'python' in the last 48h",labels={"doc_count":"# comments","key":"Subreddits"},# the axis namescolor_discrete_sequence=["blueviolet"],# the colors usedheight=500,width=800)Yes, perhaps a bit more verbose than matplotlib, but you get an interactive chart!
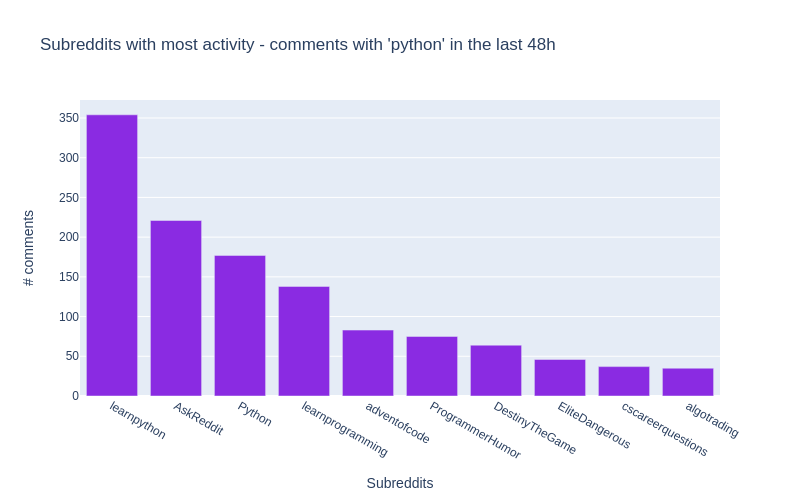
All of the details are included in the notebook for this article.
What are the most up-voted comments with the word ‘python’?
To answer this question, our function will again come in handy. Let’s aggregate things a bit.
Don’t get scared, this is a one liner that will produce similar results to above:
# get the data we need using the functiondata=get_pushshift_data(data_type="comment",q="python",after="7d",size=10,sort_type="score",sort="desc").get("data")# we only care about certain columnsdf=pandas.DataFrame.from_records(data)[["author","subreddit","score","body","permalink"]]# we only keep the first X characters of the body of the comment (sometimes they are too big)df['body']=df['body'].str[0:400]+"..."# we append the string to all the permalink entries so that we have a link to the commentdf['permalink']="https://reddit.com"+df['permalink'].astype(str)# style the last column to be clickable and printdf.style.format({'permalink':make_clickable})To make a DataFrame column clickable you can can apply the following function to it:
defmake_clickable(val):""" Makes a pandas column clickable by wrapping it in some html."""return'<a href="{}">Link</a>'.format(val,val)The above code will return the top 10 most upvoted comments of the last 7 days:
| author | subreddit | sc or e | body | permalink | |
|---|---|---|---|---|---|
| 0 | Saiboo | learnpyth on | 11 1 | Suppose you create the following python file calle… | Link |
| 1 | Kompakt | Programme rHumor | 92 | Some languages don’t have switch statements…look… | Link |
| 2 | clown_ world_ 2020 | MrRobot | 47 | Just goes to show that Esmail isn’t the only brill… | Link |
| 3 | Leebert ysauce | AnimalsBe ingBros | 28 | They wont even be mad when the python decide to ta… | Link |
| 4 | Kompakt | Programme rHumor | 23 | Yep it’s true, and depending on the design of the … | Link |
| 5 | niceboy 4431 | Cringetop ia | 23 | I have a theory (someone prove me wrong if you kno… | Link |
| 6 | kinggur u | Denmark | 22 | Brug af Python: +1 Brug af Python 3: +2… | Link |
| 7 | MintyAr oma | totalwar | 20 | We really need Bretonnian Men-At-Arms shouting Mon… | Link |
| 8 | aspirin gtobeme | gifsthatk eepongivi ng | 19 | Amazing. Brought [this Monty Python clip](… | Link |
| 9 | Crimson Spooker | TwoBestFr iendsPlay | 19 | “Why can’t Three Houses be gritty and “realistic” … | Link |
In the notebook, you can click the link column to be taken right into the comment. Hooray! 
What is the sentiment in /r/python across time? Introducing TextBlob
Alright, the final analysis is a bit more complicated. We want to see the sentiment in the /r/python subreddit in some sort of time line.
First, we already now how to retrieve the most up voted comments of the past 2 days:
# get the data with our functiondata=get_pushshift_data(data_type="comment",after="2d",size=1000,sort_type="score",sort="desc",subreddit="python").get("data")# define a list of columns we want to keepcolumns_of_interest=["author","body","created_utc","score","permalink"]# transform the response into a dataframedf=pandas.DataFrame.from_records(data)[columns_of_interest]This gives us a pandas DataFrame with the columns specified in
columns_of_interest
. But how do we get the sentiment of
every comment?
Enter TextBlob. A simple library that makes it ridiculously easy to get the sentiment of a sentence. Textblob returns two values, the sentiment polarity (-1 is negative; 0 is neutral; and 1 is positive) and the sentiment subjectivity (0 is objective and 1 is subjective)
Here’s an example:
importtextblobsentence1="Portugal is a horrible country. People drive like crazy animals."print(textblob.TextBlob(sentence1).sentiment)# -> Sentiment(polarity=-0.8, subjectivity=0.95)# negative and subjectivesentence2="Portugal is the most beautiful country in the world because beaches face west."print(textblob.TextBlob(sentence2).sentiment)# -> Sentiment(polarity=0.675, subjectivity=0.75)# positive and less subjectiveRead more about the library here.
Now that we know how to extract sentiment from a piece of text, we can easily create some other columns for our DataFrame of comments:
# create a column with sentiment polaritydf["sentiment_polarity"]=df.apply(lambdarow:textblob.TextBlob(row["body"]).sentiment.polarity,axis=1)# create a column with sentiment subjectivitydf["sentiment_subjectivity"]=df.apply(lambdarow:textblob.TextBlob(row["body"]).sentiment.subjectivity,axis=1)# create a column with 'positive' or 'negative' depending on sentiment_polaritydf["sentiment"]=df.apply(lambdarow:"positive"ifrow["sentiment_polarity"]>=0else"negative",axis=1)# create a column with a text preview that shows the first 50 charactersdf["preview"]=df["body"].str[0:50]# take the created_utc parameter and tranform it into a datetime columndf["date"]=pandas.to_datetime(df['created_utc'],unit='s')Finally, it’s time to plot our figure with the help of Plotly Express:
px.scatter(df,x="date",# date on the x axisy="sentiment_polarity",# sentiment on the y axishover_data=["author","permalink","preview"],# data to show on hovercolor_discrete_sequence=["lightseagreen","indianred"],# colors to usecolor="sentiment",# what should the color depend on?size="score",# the more votes, the bigger the circlesize_max=10,# not too biglabels={"sentiment_polarity":"Comment positivity","date":"Date comment was posted"},# axis namestitle=f"Comment sentiment in /r/python for the past 48h",# title of figure)And here’s the output!
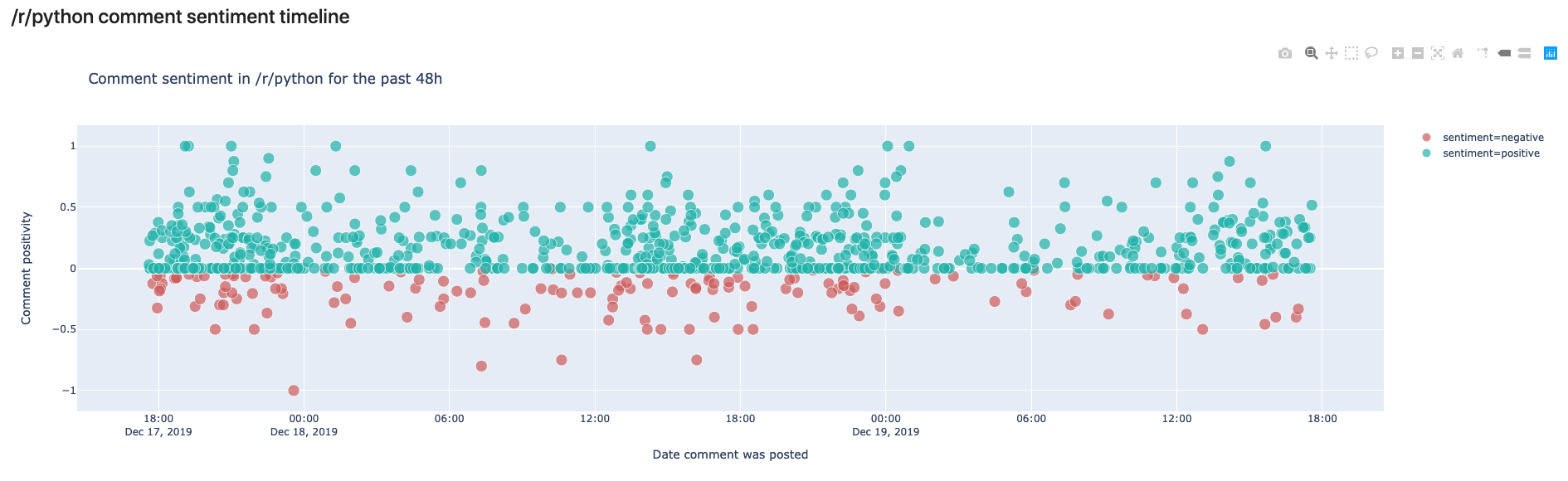
In this view, we can see the comments made in /r/python in the last 48 hours. We can see that most comments are rather on the positive side, but some are also negative. In your own notebook you’ll notice that you can hover over the comments and read the preview to see why they were classified as negative or positive.
The cool thing here is that if you run the same script tomorrow, you’ll get a different output.
So how can we have this in some place that “automatically” is updated whenever we see it?
Creating a live dashboard with Voilà
Voilà has a simple premise: “Voilà turns Jupyter notebooks into standalone web applications.”
Let’s back up a bit, and get everything you need running in your system. First step is to have a working setup with everything above, for that, follow these instructions .
Once that is done, you should be able to launch the dashboard with:
(env) $ voila notebooks/Dashboard.ipynb
Now, you should be able to see a web like application in a new tab in your browser from the notebook we created!
Feel free to modify this notebook according to your interests. You’ll notice that I have created some general variables in the first notebook cell, so you can fire up Jupyter Lab, and modify them and see what comes out!
Here are the general modifiable cells:
COMMENT_COLOR="blueviolet"# color for your comment graphSUBMISSION_COLOR="darkorange"# color for your submission graphTEXT_PREVIEW_SIZE=240# how long should the preview be?TERM_OF_INTEREST="python"# maybe you are interested in some other term?SUBREDDIT_OF_INTEREST="python"# maybe you are interested in some other subreddit?TIMEFRAME="48h"# you can define another timelineOnce you have modified your dashboard, you can launch Voilà again to see the results.
The most important thing about Voilà is that every time it runs, it actually re-runs your whole code, which yes, makes things a bit slow, but also means that the results get updated every time the page is refreshed! :tada:
Deploying your notebook to the web
First option: Using binder
Binder helps you turn a simple GitHub repo into an interactive notebook environment. They do this by using docker images to reproduce your GitHub repo’s setup.
We don’t really care about all that. We just want to publish our Voilà dashboard. To do that, follow these steps:
- Create a public GitHub repo
- Add the notebooks you want to publish as dashboards to it
- Add a
requirements.txtfile just as I have in the example repo with all of your dependencies. - Go to mybinder.org
- In the
GitHubfield add your repo’s URL. - In the
GitHub branch, tag, or commitfield, add “master”, otherwise, you probably know what you are doing. - In the
Path to a notebook fieldadd/voila/render/path/to/notebook.ipynbthepath/to/rendershould be the location of your notebook in your repo. In the example, this results invoila/render/notebooks/Dashboard.ipynb - In the
Path to a notebook fieldtoggleURL(instead of the defaultfileoption) - Hit
launch - Your dashboard will automatically launch :open_mouth: :tada:
- You can share the link with others and they will have access to the dashboard as well.
Here is the running example of our reddit dashboard. (Takes a bit to build for the first time..)
Second Option: Using an ubuntu server in a hacky way with tmux ![🙈]()
WARNING: This option is not 100% safe, so make sure to only use it for testing or proof of concepts, particularly if you are dealing with sensitive data!
If you want to have your dashboard running on a typical URL (such as mycooldash.com for example), you probably want to deploy it on a Linux server.
Here are the steps I used to accomplish that:
- Set up your virtual private server - this Linode guide is a good start.
- Make sure port 80 (the regular http port) is open
$ sudo iptables -A INPUT -p tcp --dport 80 -j ACCEPT
- Once you have your repo in GitHub or somewhere else, clone it to your server.
$ git clone https://github.com/your_username/your_awesome_repo.git
- You should already have python 3 installed. Try typing
python3in your console. If that fails then these instructions will help you. - Make sure you can run your dashboard, by creating a virtual environment and installing the dependencies.
- Now, if you type in your console the Voilà command, and specify the port:
(env) $ voila YourNoteBook.ipynb --port=80
You can probably navigate to your server’s IP and see the dashboard. However, as soon as you exit your server, your dashboard will stop working. We are going to use a nifty trick with a tool called tmux.
Tmux is a “terminal multiplexer” (wow, that’s a big word). It basically allows us to create multiple terminal sessions at the same time, and then (yes you guessed it), keep them running indefinitely. If this sounds confusing, let’s just get to it.
- Install tmux:
$ sudo apt-get install tmux
- Once installed we create a new terminal session:
$ tmux new voila
- You are now inside a new terminal session. Let’s get Voilà running there.
$ cd my_repo # navigate to the repo $ . env/bin/activate # activate the environment (env) $ voila MyNotebook.ipynb --port=80# start the dashboard on port 80
- You should see the dashboard in your browser
- And now, for the magic, in your terminal hit
ctrl+band thendon your keyboard. This will “detach” you from that terminal where Voilà is running. - You are now back to your original terminal session. Notice that your dashboard is still running. This is because your
voilaterminal session is still running. - You can see it by listing the terminal sessions with:
$ tmux ls
- And then attach to it via:
$ tmux attach voila
- And you’ll see your Voilà logs outputting.
This is arguably a bit of a hack to have things running, but it works - so no complaints there.
Tmux is an awesome tool, and you should definitely learn more about it here.
Using Heroku or Google Cloud Platform
There are a million ways of deploying, and Voilà also has good documentation on these.
Conclusion
That was a long post! But we are finally done! Let’s summarize everything we learned:
- We learned how to transform an API endpoint into a function with
*kwargs - We learned how to analyze reddit data with python and Plotly Express
- We learned how to analyze sentiment from sentences with TextBlob
- We learned how to transform a jupyter notebook into a dashboard using Voilà.
- We learned how to deploy those dashboards with Binder.org
- We learned how to use tmux to deploy these kinds of tools in a server.
That was a lot of stuff, and probably there are a lot of bugs in my notebook, or explanation so make sure to:
- Visit the GitHub repo were both the code and post are stored.
- If there is something wrong in the code please feel free to submit an issue or a pull request.
- Tweet at me if you have questions!
- Visit my website if you want to learn more about my work
![😀]()
Hope you enjoyed it!
Real Python: NumPy, SciPy, and Pandas: Correlation With Python
Correlation coefficients quantify the association between variables or features of a dataset. These statistics are of high importance for science and technology, and Python has great tools that you can use to calculate them. SciPy, NumPy, and Pandas correlation methods are fast, comprehensive, and well-documented.
In this tutorial, you’ll learn:
- What Pearson, Spearman, and Kendall correlation coefficients are
- How to use SciPy, NumPy, and Pandas correlation functions
- How to visualize data, regression lines, and correlation matrices with Matplotlib
You’ll start with an explanation of correlation, then see three quick introductory examples, and finally dive into details of NumPy, SciPy and Pandas correlation.
Free Bonus:Click here to get access to a free NumPy Resources Guide that points you to the best tutorials, videos, and books for improving your NumPy skills.
Correlation
Statistics and data science are often concerned about the relationships between two or more variables (or features) of a dataset. Each data point in the dataset is an observation, and the features are the properties or attributes of those observations.
Every dataset you work with uses variables and observations. For example, you might be interested in understanding the following:
- How the height of basketball players is correlated to their shooting accuracy
- Whether there’s a relationship between employee work experience and salary
- What mathematical dependence exists between the population density and the gross domestic product of different countries
In the examples above, the height, shooting accuracy, years of experience, salary, population density, and gross domestic product are the features or variables. The data related to each player, employee, and each country are the observations.
When data is represented in the form of a table, the rows of that table are usually the observations, while the columns are the features. Take a look at this employee table:
| Name | Years of Experience | Annual Salary |
|---|---|---|
| Ann | 30 | 120,000 |
| Rob | 21 | 105,000 |
| Tom | 19 | 90,000 |
| Ivy | 10 | 82,000 |
In this table, each row represents one observation, or the data about one employee (either Ann, Rob, Tom, or Ivy). Each column shows one property or feature (name, experience, or salary) for all the employees.
If you analyze any two features of a dataset, then you’ll find some type of correlation between those two features. Consider the following figures:
Each of these plots shows one of three different forms of correlation:
Negative correlation (red dots): In the plot on the left, the y values tend to decrease as the x values increase. This shows strong negative correlation, which occurs when large values of one feature correspond to small values of the other, and vice versa.
Weak or no correlation (green dots): The plot in the middle shows no obvious trend. This is a form of weak correlation, which occurs when an association between two features is not obvious or is hardly observable.
Positive correlation (blue dots): In the plot on the right, the y values tend to increase as the x values increase. This illustrates strong positive correlation, which occurs when large values of one feature correspond to large values of the other, and vice versa.
The next figure represents the data from the employee table above:
The correlation between experience and salary is positive because higher experience corresponds to a larger salary and vice versa.
Note: When you’re analyzing correlation, you should always have in mind that correlation does not indicate causation. It quantifies the strength of the relationship between the features of a dataset. Sometimes, the association is caused by a factor common to several features of interest.
Correlation is tightly connected to other statistical quantities like the mean, standard deviation, variance, and covariance. If you want to learn more about these quantities and how to calculate them with Python, then check out Descriptive Statistics with Python.
There are several statistics that you can use to quantify correlation. In this tutorial, you’ll learn about three correlation coefficients:
Pearson’s coefficient measures linear correlation, while the Spearman and Kendall coefficients compare the ranks of data. There are several NumPy, SciPy, and Pandas correlation functions and methods that you can use to calculate these coefficients. You can also use Matplotlib to conveniently illustrate the results.
Example: NumPy Correlation Calculation
NumPy has many statistics routines, including np.corrcoef(), that return a matrix of Pearson correlation coefficients. You can start by importing NumPy and defining two NumPy arrays. These are instances of the class ndarray. Call them x and y:
>>> importnumpyasnp>>> x=np.arange(10,20)>>> xarray([10, 11, 12, 13, 14, 15, 16, 17, 18, 19])>>> y=np.array([2,1,4,5,8,12,18,25,96,48])>>> yarray([ 2, 1, 4, 5, 8, 12, 18, 25, 96, 48])Here, you use np.arange() to create an array x of integers between 10 (inclusive) and 20 (exclusive). Then you use np.array() to create a second array y containing arbitrary integers.
Once you have two arrays of the same length, you can call np.corrcoef() with both arrays as arguments:
>>> r=np.corrcoef(x,y)>>> rarray([[1. , 0.75864029], [0.75864029, 1. ]])>>> r[0,1]0.7586402890911867>>> r[1,0]0.7586402890911869corrcoef() returns the correlation matrix, which is a two-dimensional array with the correlation coefficients. Here’s a simplified version of the correlation matrix you just created:
x y
x 1.00 0.76
y 0.76 1.00
The values on the main diagonal of the correlation matrix (upper left and lower right) are equal to 1. The upper left value corresponds to the correlation coefficient for x and x, while the lower right value is the correlation coefficient for y and y. They are always equal to 1.
However, what you usually need are the lower left and upper right values of the correlation matrix. These values are equal and both represent the Pearson correlation coefficient for x and y. In this case, it’s approximately 0.76.
This figure shows the data points and the correlation coefficients for the above example:
The red squares are the data points. As you can see, the figure also shows the values of the three correlation coefficients.
Example: SciPy Correlation Calculation
SciPy also has many statistics routines contained in scipy.stats. You can use the following methods to calculate the three correlation coefficients you saw earlier:
Here’s how you would use these functions in Python:
>>> importnumpyasnp>>> importscipy.stats>>> x=np.arange(10,20)>>> y=np.array([2,1,4,5,8,12,18,25,96,48])>>> scipy.stats.pearsonr(x,y)# Pearson's r(0.7586402890911869, 0.010964341301680832)>>> scipy.stats.spearmanr(x,y)# Spearman's rhoSpearmanrResult(correlation=0.9757575757575757, pvalue=1.4675461874042197e-06)>>> scipy.stats.kendalltau(x,y)# Kendall's tauKendalltauResult(correlation=0.911111111111111, pvalue=2.9761904761904762e-05)Note that these functions return objects that contain two values:
- The correlation coefficient
- The p-value
You use the p-value in statistical methods when you’re testing a hypothesis. The p-value is an important measure that requires in-depth knowledge of probability and statistics to interpret. To learn more about them, you can read about the basics or check out a data scientist’s explanation of p-values.
You can extract the p-values and the correlation coefficients with their indices, as the items of tuples:
>>> scipy.stats.pearsonr(x,y)[0]# Pearson's r0.7586402890911869>>> scipy.stats.spearmanr(x,y)[0]# Spearman's rho0.9757575757575757>>> scipy.stats.kendalltau(x,y)[0]# Kendall's tau0.911111111111111You could also use dot notation for the Spearman and Kendall coefficients:
>>> scipy.stats.spearmanr(x,y).correlation# Spearman's rho0.9757575757575757>>> scipy.stats.kendalltau(x,y).correlation# Kendall's tau0.911111111111111The dot notation is longer, but it’s also more readable and more self-explanatory.
If you want to get the Pearson correlation coefficient and p-value at the same time, then you can unpack the return value:
>>> r,p=scipy.stats.pearsonr(x,y)>>> r0.7586402890911869>>> p0.010964341301680829This approach exploits Python unpacking and the fact that pearsonr() returns a tuple with these two statistics. You can also use this technique with spearmanr() and kendalltau(), as you’ll see later on.
Example: Pandas Correlation Calculation
Pandas is, in some cases, more convenient than NumPy and SciPy for calculating statistics. It offers statistical methods for Series and DataFrame instances. For example, given two Series objects with the same number of items, you can call .corr() on one of them with the other as the first argument:
>>> importpandasaspd>>> x=pd.Series(range(10,20))>>> x0 101 112 123 134 145 156 167 178 189 19dtype: int64>>> y=pd.Series([2,1,4,5,8,12,18,25,96,48])>>> y0 21 12 43 54 85 126 187 258 969 48dtype: int64>>> x.corr(y)# Pearson's r0.7586402890911867>>> y.corr(x)0.7586402890911869>>> x.corr(y,method='spearman')# Spearman's rho0.9757575757575757>>> x.corr(y,method='kendall')# Kendall's tau0.911111111111111Here, you use .corr() to calculate all three correlation coefficients. You define the desired statistic with the parameter method, which can take on one of several values:
'pearson''spearman''kendall'- a callable
The callable can be any function, method, or object with .__call__() that accepts two one-dimensional arrays and returns a floating-point number.
Linear Correlation
Linear correlation measures the proximity of the mathematical relationship between variables or dataset features to a linear function. If the relationship between the two features is closer to some linear function, then their linear correlation is stronger and the absolute value of the correlation coefficient is higher.
Pearson Correlation Coefficient
Consider a dataset with two features: x and y. Each feature has n values, so x and y are n-tuples. Say that the first value x₁ from x corresponds to the first value y₁ from y, the second value x₂ from x to the second value y₂ from y, and so on. Then, there are n pairs of corresponding values: (x₁, y₁), (x₂, y₂), and so on. Each of these x-y pairs represents a single observation.
The Pearson (product-moment) correlation coefficient is a measure of the linear relationship between two features. It’s the ratio of the covariance of x and y to the product of their standard deviations. It’s often denoted with the letter r and called Pearson’s r. You can express this value mathematically with this equation:
r = Σᵢ((xᵢ − mean(x))(yᵢ − mean(y))) (√Σᵢ(xᵢ − mean(x))² √Σᵢ(yᵢ − mean(y))²)⁻¹
Here, i takes on the values 1, 2, …, n. The mean values of x and y are denoted with mean(x) and mean(y). This formula shows that if larger x values tend to correspond to larger y values and vice versa, then r is positive. On the other hand, if larger x values are mostly associated with smaller y values and vice versa, then r is negative.
Here are some important facts about the Pearson correlation coefficient:
The Pearson correlation coefficient can take on any real value in the range −1 ≤ r ≤ 1.
The maximum value r = 1 corresponds to the case when there’s a perfect positive linear relationship between x and y. In other words, larger x values correspond to larger y values and vice versa.
The value r > 0 indicates positive correlation between x and y.
The value r = 0 corresponds to the case when x and y are independent.
The value r < 0 indicates negative correlation between x and y.
The minimal value r = −1 corresponds to the case when there’s a perfect negative linear relationship between x and y. In other words, larger x values correspond to smaller y values and vice versa.
The above facts can be summed up in the following table:
| Pearson’s r Value | Correlation Between x and y |
|---|---|
| equal to 1 | perfect positive linear relationship |
| greater than 0 | positive correlation |
| equal to 0 | independent |
| less than 0 | negative correlation |
| equal to -1 | perfect negative linear relationship |
In short, a larger absolute value of r indicates stronger correlation, closer to a linear function. A smaller absolute value of r indicates weaker correlation.
Linear Regression: SciPy Implementation
Linear regression is the process of finding the linear function that is as close as possible to the actual relationship between features. In other words, you determine the linear function that best describes the association between the features. This linear function is also called the regression line.
You can implement linear regression with SciPy. You’ll get the linear function that best approximates the relationship between two arrays, as well as the Pearson correlation coefficient. To get started, you first need to import the libraries and prepare some data to work with:
>>> importnumpyasnp>>> importscipy.stats>>> x=np.arange(10,20)>>> y=np.array([2,1,4,5,8,12,18,25,96,48])Here, you import numpy and scipy.stats and define the variables x and y.
You can use scipy.stats.linregress() to perform linear regression for two arrays of the same length. You should provide the arrays as the arguments and get the outputs by using dot notation:
>>> result=scipy.stats.linregress(x,y)>>> result.slope7.4363636363636365>>> result.intercept-85.92727272727274>>> result.rvalue0.7586402890911869>>> result.pvalue0.010964341301680825>>> result.stderr2.257878767543913That’s it! You’ve completed the linear regression and gotten the following results:
.slope: the slope of the regression line.intercept: the intercept of the regression line.pvalue: the p-value.stderr: the standard error of the estimated gradient
You’ll learn how to visualize these results in a later section.
You can also provide a single argument to linregress(), but it must be a two-dimensional array with one dimension of length two:
>>> xy=np.array([[10,11,12,13,14,15,16,17,18,19],... [2,1,4,5,8,12,18,25,96,48]])>>> scipy.stats.linregress(xy)LinregressResult(slope=7.4363636363636365, intercept=-85.92727272727274, rvalue=0.7586402890911869, pvalue=0.010964341301680825, stderr=2.257878767543913)The result is exactly the same as the previous example because xy contains the same data as x and y together. linregress() took the first row of xy as one feature and the second row as the other feature.
Note: In the example above, scipy.stats.linregress() considers the rows as features and columns as observations. That’s because there are two rows.
The usual practice in machine learning is the opposite: rows are observations and columns are features. Many machine learning libraries, like Pandas, Scikit-Learn, Keras, and others, follow this convention.
You should be careful to note how the observations and features are indicated whenever you’re analyzing correlation in a dataset.
linregress() will return the same result if you provide the transpose of xy, or a NumPy array with 10 rows and two columns. In NumPy, you can transpose a matrix in many ways:
Here’s how you might transpose xy:
>>> xy.Tarray([[10, 2], [11, 1], [12, 4], [13, 5], [14, 8], [15, 12], [16, 18], [17, 25], [18, 96], [19, 48]])Now that you know how to get the transpose, you can pass one to linregress(). The first column will be one feature and the second column the other feature:
>>> scipy.stats.linregress(xy.T)LinregressResult(slope=7.4363636363636365, intercept=-85.92727272727274, rvalue=0.7586402890911869, pvalue=0.010964341301680825, stderr=2.257878767543913)Here, you use .T to get the transpose of xy. linregress() works the same way with xy and its transpose. It extracts the features by splitting the array along the dimension with length two.
You should also be careful to note whether or not your dataset contains missing values. In data science and machine learning, you’ll often find some missing or corrupted data. The usual way to represent it in Python, NumPy, SciPy, and Pandas is by using NaN or Not a Number values. But if your data contains nan values, then you won’t get a useful result with linregress():
>>> scipy.stats.linregress(np.arange(3),np.array([2,np.nan,5]))LinregressResult(slope=nan, intercept=nan, rvalue=nan, pvalue=nan, stderr=nan)In this case, your resulting object returns all nan values. In Python, nan is a special floating-point value that you can get by using any of the following:
You can also check whether a variable corresponds to nan with math.isnan() or numpy.isnan().
Pearson Correlation: NumPy and SciPy Implementation
You’ve already seen how to get the Pearson correlation coefficient with corrcoef() and pearsonr():
>>> r,p=scipy.stats.pearsonr(x,y)>>> r0.7586402890911869>>> p0.010964341301680829>>> np.corrcoef(x,y)array([[1. , 0.75864029], [0.75864029, 1. ]])Note that if you provide an array with a nan value to pearsonr(), you’ll get a ValueError.
There are few additional details worth considering. First, recall that np.corrcoef() can take two NumPy arrays as arguments. Instead, you can pass a single two-dimensional array with the same values as the argument:
>>> np.corrcoef(xy)array([[1. , 0.75864029], [0.75864029, 1. ]])The results are the same in this and previous examples. Again, the first row of xy represents one feature, while the second row represents the other.
If you want to get the correlation coefficients for three features, then you just provide a numeric two-dimensional array with three rows as the argument:
>>> xyz=np.array([[10,11,12,13,14,15,16,17,18,19],... [2,1,4,5,8,12,18,25,96,48],... [5,3,2,1,0,-2,-8,-11,-15,-16]])>>> np.corrcoef(xyz)array([[ 1. , 0.75864029, -0.96807242], [ 0.75864029, 1. , -0.83407922], [-0.96807242, -0.83407922, 1. ]])You’ll obtain the correlation matrix again, but this one will be larger than previous ones:
x y z
x 1.00 0.76 -0.97
y 0.76 1.00 -0.83
z -0.97 -0.83 1.00
This is because corrcoef() considers each row of xyz as one feature. The value 0.76 is the correlation coefficient for the first two features of xyz. This is the same as the coefficient for x and y in previous examples. -0.97 represents Pearson’s r for the first and third features, while -0.83 is Pearson’s r for the last two features.
Here’s an interesting example of what happens when you pass nan data to corrcoef():
>>> arr_with_nan=np.array([[0,1,2,3],... [2,4,1,8],... [2,5,np.nan,2]])>>> np.corrcoef(arr_with_nan)array([[1. , 0.62554324, nan], [0.62554324, 1. , nan], [ nan, nan, nan]])In this example, the first two rows (or features) of arr_with_nan are okay, but the third row [2, 5, np.nan, 2] contains a nan value. Everything that doesn’t include the feature with nan is calculated well. The results that depend on the last row, however, are nan.
By default, numpy.corrcoef() considers the rows as features and the columns as observations. If you want the opposite behavior, which is widely used in machine learning, then use the argument rowvar=False:
>>> xyz.Tarray([[ 10, 2, 5], [ 11, 1, 3], [ 12, 4, 2], [ 13, 5, 1], [ 14, 8, 0], [ 15, 12, -2], [ 16, 18, -8], [ 17, 25, -11], [ 18, 96, -15], [ 19, 48, -16]])>>> np.corrcoef(xyz.T,rowvar=False)array([[ 1. , 0.75864029, -0.96807242], [ 0.75864029, 1. , -0.83407922], [-0.96807242, -0.83407922, 1. ]])This array is identical to the one you saw earlier. Here, you apply a different convention, but the result is the same.
Pearson Correlation: Pandas Implementation
So far, you’ve used Series and DataFrame object methods to calculate correlation coefficients. Let’s explore these methods in more detail. First, you need to import Pandas and create some instances of Series and DataFrame:
>>> importpandasaspd>>> x=pd.Series(range(10,20))>>> x0 101 112 123 134 145 156 167 178 189 19dtype: int64>>> y=pd.Series([2,1,4,5,8,12,18,25,96,48])>>> y0 21 12 43 54 85 126 187 258 969 48dtype: int64>>> z=pd.Series([5,3,2,1,0,-2,-8,-11,-15,-16])>>> z0 51 32 23 14 05 -26 -87 -118 -159 -16dtype: int64>>> xy=pd.DataFrame({'x-values':x,'y-values':y})>>> xy x-values y-values0 10 21 11 12 12 43 13 54 14 85 15 126 16 187 17 258 18 969 19 48>>> xyz=pd.DataFrame({'x-values':x,'y-values':y,'z-values':z})>>> xyz x-values y-values z-values0 10 2 51 11 1 32 12 4 23 13 5 14 14 8 05 15 12 -26 16 18 -87 17 25 -118 18 96 -159 19 48 -16You now have three Series objects called x, y, and z. You also have two DataFrame objects, xy and xyz.
Note: When you work with DataFrame instances, you should be aware that the rows are observations and the columns are features. This is consistent with the usual practice in machine learning.
You’ve already learned how to use .corr() with Series objects to get the Pearson correlation coefficient:
>>> x.corr(y)0.7586402890911867Here, you call .corr() on one object and pass the other as the first argument.
If you provide a nan value, then .corr() will still work, but it will exclude observations that contain nan values:
>>> u,u_with_nan=pd.Series([1,2,3]),pd.Series([1,2,np.nan,3])>>> v,w=pd.Series([1,4,8]),pd.Series([1,4,154,8])>>> u.corr(v)0.9966158955401239>>> u_with_nan.corr(w)0.9966158955401239You get the same value of the correlation coefficient in these two examples. That’s because .corr() ignores the pair of values (np.nan, 154) that has a missing value.
You can also use .corr() with DataFrame objects. You can use it to get the correlation matrix for their columns:
>>> corr_matrix=xy.corr()>>> corr_matrix x-values y-valuesx-values 1.00000 0.75864y-values 0.75864 1.00000The resulting correlation matrix is a new instance of DataFrame and holds the correlation coefficients for the columns xy['x-values'] and xy['y-values']. Such labeled results are usually very convenient to work with because you can access them with either their labels or their integer position indices:
>>> corr_matrix.at['x-values','y-values']0.7586402890911869>>> corr_matrix.iat[0,1]0.7586402890911869This example shows two ways of accessing values:
- Use
.at[]to access a single value by row and column labels. - Use
.iat[]to access a value by the positions of its row and column.
You can apply .corr() the same way with DataFrame objects that contain three or more columns:
>>> xyz.corr() x-values y-values z-valuesx-values 1.000000 0.758640 -0.968072y-values 0.758640 1.000000 -0.834079z-values -0.968072 -0.834079 1.000000You’ll get a correlation matrix with the following correlation coefficients:
0.758640forx-valuesandy-values-0.968072forx-valuesandz-values-0.834079fory-valuesandz-values
Another useful method is .corrwith(), which allows you to calculate the correlation coefficients between the rows or columns of one DataFrame object and another Series or DataFrame object passed as the first argument:
>>> xy.corrwith(z)x-values -0.968072y-values -0.834079dtype: float64In this case, the result is a new Series object with the correlation coefficient for the column xy['x-values'] and the values of z, as well as the coefficient for xy['y-values'] and z.
.corrwith() has the optional parameter axis that specifies whether columns or rows represent the features. The default value of axis is 0, and it also defaults to columns representing features. There’s also a drop parameter, which indicates what to do with missing values.
Both .corr() and .corrwith() have the optional parameter method to specify the correlation coefficient that you want to calculate. The Pearson correlation coefficient is returned by default, so you don’t need to provide it in this case.
Rank Correlation
Rank correlation compares the ranks or the orderings of the data related to two variables or dataset features. If the orderings are similar, then the correlation is strong, positive, and high. However, if the orderings are close to reversed, then the correlation is strong, negative, and low. In other words, rank correlation is concerned only with the order of values, not with the particular values from the dataset.
To illustrate the difference between linear and rank correlation, consider the following figure:
The left plot has a perfect positive linear relationship between x and y, so r = 1. The central plot shows positive correlation and the right one shows negative correlation. However, neither of them is a linear function, so r is different than −1 or 1.
When you look only at the orderings or ranks, all three relationships are perfect! The left and central plots show the observations where larger x values always correspond to larger y values. This is perfect positive rank correlation. The right plot illustrates the opposite case, which is perfect negative rank correlation.
Spearman Correlation Coefficient
The Spearman correlation coefficient between two features is the Pearson correlation coefficient between their rank values. It’s calculated the same way as the Pearson correlation coefficient but takes into account their ranks instead of their values. It’s often denoted with the Greek letter rho (ρ) and called Spearman’s rho.
Say you have two n-tuples, x and y, where (x₁, y₁), (x₂, y₂), … are the observations as pairs of corresponding values. You can calculate the Spearman correlation coefficient ρ the same way as the Pearson coefficient. You’ll use the ranks instead of the actual values from x and y.
Here are some important facts about the Pearson correlation coefficient:
It can take a real value in the range −1 ≤ ρ ≤ 1.
Its maximum value ρ = 1 corresponds to the case when there’s a monotonically increasing function between x and y. In other words, larger x values correspond to larger y values and vice versa.
Its minimum value ρ = −1 corresponds to the case when there’s a monotonically decreasing function between x and y. In other words, larger x values correspond to smaller y values and vice versa.
You can calculate Spearman’s rho in Python in a very similar way as you would Pearson’s r.
Kendall Correlation Coefficient
Let’s start again by considering two n-tuples, x and y. Each of the x-y pairs (x₁, y₁), (x₂, y₂), … is a single observation. A pair of observations (xᵢ, yᵢ) and (xⱼ, yⱼ), where i < j, will be one of three things:
- concordant if either (xᵢ > xⱼ and yᵢ > yⱼ) or (xᵢ < xⱼ and yᵢ < yⱼ)
- discordant if either (xᵢ < xⱼ and yᵢ > yⱼ) or (xᵢ > xⱼ and yᵢ < yⱼ)
- neither if there’s a tie in x (xᵢ = xⱼ) or a tie in y (yᵢ = yⱼ)
The Kendall correlation coefficient compares the number of concordant and discordant pairs of data. This coefficient is based on the difference in the counts of concordant and discordant pairs relative to the number of x-y pairs. It’s often denoted with the Greek letter tau (τ) and called Kendall’s tau.
According to the scipy.stats official docs, the Kendall correlation coefficient is calculated as
τ = (n⁺ − n⁻) / √((n⁺ + n⁻ + nˣ)(n⁺ + n⁻ + nʸ)),
where:
- n⁺ is the number of concordant pairs
- n⁻ is the number of discordant pairs
- nˣ is the number of ties only in x
- nʸ is the number of ties only in y
If a tie occurs in both x and y, then it’s not included in either nˣ or nʸ.
The Wikipedia page on Kendall rank correlation coefficient gives the following expression: τ = (2 / (n(n − 1))) Σᵢⱼ(sign(xᵢ − xⱼ) sign(yᵢ − yⱼ)) for i < j, where i = 1, 2, …, n − 1 and j = 2, 3, …, n. The sign function sign(z) is −1 if z < 0, 0 if z = 0, and 1 if z > 0. n(n − 1) / 2 is the total number of x-y pairs.
Some important facts about the Kendall correlation coefficient are as follows:
It can take a real value in the range −1 ≤ τ ≤ 1.
Its maximum value τ = 1 corresponds to the case when the ranks of the corresponding values in x and y are the same. In other words, all pairs are concordant.
Its minimum value τ = −1 corresponds to the case when the rankings in x are the reverse of the rankings in y. In other words, all pairs are discordant.
You can calculate Kendall’s tau in Python similarly to how you would calculate Pearson’s r.
Rank: SciPy Implementation
You can use scipy.stats to determine the rank for each value in an array. First, you’ll import the libraries and create NumPy arrays:
>>> importnumpyasnp>>> importscipy.stats>>> x=np.arange(10,20)>>> y=np.array([2,1,4,5,8,12,18,25,96,48])>>> z=np.array([5,3,2,1,0,-2,-8,-11,-15,-16])Now that you’ve prepared data, you can determine the rank of each value in a NumPy array with scipy.stats.rankdata():
>>> scipy.stats.rankdata(x)array([ 1., 2., 3., 4., 5., 6., 7., 8., 9., 10.])>>> scipy.stats.rankdata(y)array([ 2., 1., 3., 4., 5., 6., 7., 8., 10., 9.])>>> scipy.stats.rankdata(z)array([10., 9., 8., 7., 6., 5., 4., 3., 2., 1.])The arrays x and z are monotonic, so their ranks are monotonic as well. The smallest value in y is 1 and it corresponds to the rank 1. The second smallest is 2, which corresponds to the rank 2. The largest value is 96, which corresponds to the largest rank 10 since there are 10 items in the array.
rankdata() has the optional parameter method. This tells Python what to do if there are ties in the array (if two or more values are equal). By default, it assigns them the average of the ranks:
>>> scipy.stats.rankdata([8,2,0,2])array([4. , 2.5, 1. , 2.5])There are two elements with a value of 2 and they have the ranks 2.0 and 3.0. The value 0 has rank 1.0 and the value 8 has rank 4.0. Then, both elements with the value 2 will get the same rank 2.5.
rankdata() treats nan values as if they were large:
>>> scipy.stats.rankdata([8,np.nan,0,2])array([3., 4., 1., 2.])In this case, the value np.nan corresponds to the largest rank 4.0. You can also get ranks with np.argsort():
>>> np.argsort(y)+1array([ 2, 1, 3, 4, 5, 6, 7, 8, 10, 9])argsort() returns the indices that the array items would have in the sorted array. These indices are zero-based, so you’ll need to add 1 to all of them.
Rank Correlation: NumPy and SciPy Implementation
You can calculate the Spearman correlation coefficient with scipy.stats.spearmanr():
>>> result=scipy.stats.spearmanr(x,y)>>> resultSpearmanrResult(correlation=0.9757575757575757, pvalue=1.4675461874042197e-06)>>> result.correlation0.9757575757575757>>> result.pvalue1.4675461874042197e-06>>> rho,p=scipy.stats.spearmanr(x,y)>>> rho0.9757575757575757>>> p1.4675461874042197e-06spearmanr() returns an object that contains the value of the Spearman correlation coefficient and p-value. As you can see, you can access particular values in two ways:
- Using dot notation (
result.correlationandresult.pvalue) - Using Python unpacking (
rho, p = scipy.stats.spearmanr(x, y))
You can get the same result if you provide the two-dimensional array xy that contains the same data as x and y to spearmanr():
>>> xy=np.array([[10,11,12,13,14,15,16,17,18,19],... [2,1,4,5,8,12,18,25,96,48]])>>> rho,p=scipy.stats.spearmanr(xy,axis=1)>>> rho0.9757575757575757>>> p1.4675461874042197e-06The first row of xy is one feature, while the second row is the other feature. You can modify this. The optional parameter axis determines whether columns (axis=0) or rows (axis=1) represent the features. The default behavior is that the rows are observations and the columns are features.
Another optional parameter nan_policy defines how to handle nan values. It can take one of three values:
'propagate'returnsnanif there’s ananvalue among the inputs. This is the default behavior.'raise'raises aValueErrorif there’s ananvalue among the inputs.'omit'ignores the observations withnanvalues.
If you provide a two-dimensional array with more than two features, then you’ll get the correlation matrix and the matrix of the p-values:
>>> xyz=np.array([[10,11,12,13,14,15,16,17,18,19],... [2,1,4,5,8,12,18,25,96,48],... [5,3,2,1,0,-2,-8,-11,-15,-16]])>>> corr_matrix,p_matrix=scipy.stats.spearmanr(xyz,axis=1)>>> corr_matrixarray([[ 1. , 0.97575758, -1. ], [ 0.97575758, 1. , -0.97575758], [-1. , -0.97575758, 1. ]])>>> p_matrixarray([[6.64689742e-64, 1.46754619e-06, 6.64689742e-64], [1.46754619e-06, 6.64689742e-64, 1.46754619e-06], [6.64689742e-64, 1.46754619e-06, 6.64689742e-64]])The value -1 in the correlation matrix shows that the first and third features have a perfect negative rank correlation, that is that larger values in the first row always correspond to smaller values in the third.
You can obtain the Kendall correlation coefficient with kendalltau():
>>> result=scipy.stats.kendalltau(x,y)>>> resultKendalltauResult(correlation=0.911111111111111, pvalue=2.9761904761904762e-05)>>> result.correlation0.911111111111111>>> result.pvalue2.9761904761904762e-05>>> tau,p=scipy.stats.kendalltau(x,y)>>> tau0.911111111111111>>> p2.9761904761904762e-05kendalltau() works much like spearmanr(). It takes two one-dimensional arrays, has the optional parameter nan_policy, and returns an object with the values of the correlation coefficient and p-value.
However, if you provide only one two-dimensional array as an argument, then kendalltau() will raise a TypeError. If you pass two multi-dimensional arrays of the same shape, then they’ll be flattened before the calculation.
Rank Correlation: Pandas Implementation
You can calculate the Spearman and Kendall correlation coefficients with Pandas. Just like before, you start by importing pandas and creating some Series and DataFrame instances:
>>> importpandasaspd>>> x,y,z=pd.Series(x),pd.Series(y),pd.Series(z)>>> xy=pd.DataFrame({'x-values':x,'y-values':y})>>> xyz=pd.DataFrame({'x-values':x,'y-values':y,'z-values':z})Now that you have these Pandas objects, you can use .corr() and .corrwith() just like you did when you calculated the Pearson correlation coefficient. You just need to specify the desired correlation coefficient with the optional parameter method, which defaults to 'pearson'.
To calculate Spearman’s rho, pass method=spearman:
>>> x.corr(y,method='spearman')0.9757575757575757>>> xy.corr(method='spearman') x-values y-valuesx-values 1.000000 0.975758y-values 0.975758 1.000000>>> xyz.corr(method='spearman') x-values y-values z-valuesx-values 1.000000 0.975758 -1.000000y-values 0.975758 1.000000 -0.975758z-values -1.000000 -0.975758 1.000000>>> xy.corrwith(z,method='spearman')x-values -1.000000y-values -0.975758dtype: float64If you want Kendall’s tau, then you use method=kendall:
>>> x.corr(y,method='kendall')0.911111111111111>>> xy.corr(method='kendall') x-values y-valuesx-values 1.000000 0.911111y-values 0.911111 1.000000>>> xyz.corr(method='kendall') x-values y-values z-valuesx-values 1.000000 0.911111 -1.000000y-values 0.911111 1.000000 -0.911111z-values -1.000000 -0.911111 1.000000>>> xy.corrwith(z,method='kendall')x-values -1.000000y-values -0.911111dtype: float64As you can see, unlike with SciPy, you can use a single two-dimensional data structure (a dataframe).
Visualization of Correlation
Data visualization is very important in statistics and data science. It can help you better understand your data and give you a better insight into the relationships between features. In this section, you’ll learn how to visually represent the relationship between two features with an x-y plot. You’ll also use heatmaps to visualize a correlation matrix.
You’ll learn how to prepare data and get certain visual representations, but you won’t cover many other explanations. To learn more about Matplotlib in-depth, check out Python Plotting With Matplotlib (Guide). You can also take a look at the official documentation and Anatomy of Matplotlib.
To get started, first import matplotlib.pyplot:
>>> importmatplotlib.pyplotasplt>>> plt.style.use('ggplot')Here, you use plt.style.use('ggplot') to set the style of the plots. Feel free to skip this line if you want.
You’ll use the arrays x, y, z, and xyz from the previous sections. You can create them again to cut down on scrolling:
>>> importnumpyasnp>>> importscipy.stats>>> x=np.arange(10,20)>>> y=np.array([2,1,4,5,8,12,18,25,96,48])>>> z=np.array([5,3,2,1,0,-2,-8,-11,-15,-16])>>> xyz=np.array([[10,11,12,13,14,15,16,17,18,19],... [2,1,4,5,8,12,18,25,96,48],... [5,3,2,1,0,-2,-8,-11,-15,-16]])Now that you’ve got your data, you’re ready to plot.
X-Y Plots With a Regression Line
First, you’ll see how to create an x-y plot with the regression line, its equation, and the Pearson correlation coefficient. You can get the slope and the intercept of the regression line, as well as the correlation coefficient, with linregress():
>>> slope,intercept,r,p,stderr=scipy.stats.linregress(x,y)Now you have all the values you need. You can also get the string with the equation of the regression line and the value of the correlation coefficient. f-strings are very convenient for this purpose:
>>> line=f'Regression line: y={intercept:.2f}+{slope:.2f}x, r={r:.2f}'>>> line'Regression line: y=-85.93+7.44x, r=0.76'Now, create the x-y plot with .plot():
fig,ax=plt.subplots()ax.plot(x,y,linewidth=0,marker='s',label='Data points')ax.plot(x,intercept+slope*x,label=line)ax.set_xlabel('x')ax.set_ylabel('y')ax.legend(facecolor='white')plt.show()Your output should look like this:
The red squares represent the observations, while the blue line is the regression line. Its equation is listed in the legend, together with the correlation coefficient.
Heatmaps of Correlation Matrices
The correlation matrix can become really big and confusing when you have a lot of features! Fortunately, you can present it visually as a heatmap where each field has the color that corresponds to its value. You’ll need the correlation matrix:
>>> corr_matrix=np.corrcoef(xyz).round(decimals=2)>>> corr_matrixarray([[ 1. , 0.76, -0.97], [ 0.76, 1. , -0.83], [-0.97, -0.83, 1. ]])It can be convenient for you to round the numbers in the correlation matrix with .round(), as they’re going to be shown be on the heatmap.
Finally, create your heatmap with .imshow() and the correlation matrix as its argument:
fig,ax=plt.subplots()im=ax.imshow(corr_matrix)im.set_clim(-1,1)ax.grid(False)ax.xaxis.set(ticks=(0,1,2),ticklabels=('x','y','z'))ax.yaxis.set(ticks=(0,1,2),ticklabels=('x','y','z'))ax.set_ylim(2.5,-0.5)foriinrange(3):forjinrange(3):ax.text(j,i,corr_matrix[i,j],ha='center',va='center',color='r')cbar=ax.figure.colorbar(im,ax=ax,format='% .2f')plt.show()Your output should look like this:
The result is a table with the coefficients. It sort of looks like the Pandas output with colored backgrounds. The colors help you interpret the output. In this example, the yellow color represents the number 1, green corresponds to 0.76, and purple is used for the negative numbers.
Conclusion
You now know that correlation coefficients are statistics that measure the association between variables or features of datasets. They’re very important in data science and machine learning.
You can now use Python to calculate:
- Pearson’s product-moment correlation coefficient
- Spearman’s rank correlation coefficient
- Kendall’s rank correlation coefficient
Now you can use NumPy, SciPy, and Pandas correlation functions and methods to effectively calculate these (and other) statistics, even when you work with large datasets. You also know how to visualize data, regression lines, and correlation matrices with Matplotlib plots and heatmaps.
If you have any questions or comments, please put them in the comments section below!
[ Improve Your Python With 🐍 Python Tricks 💌 – Get a short & sweet Python Trick delivered to your inbox every couple of days. >> Click here to learn more and see examples ]
Codementor: Guide to the Latest Trend in Fintech Area - RegTech
Stein Magnus Jodal: 10 years of Mopidy
Ten years ago today, on December 23, 2009, Mopidy was born. While chatting with my friend and then-colleague Johannes Knutsen, we came up with the idea of building an MPD server that could play music from Spotify instead of local files.
This is the story of the first decade of Mopidy.
After a brief discussion of how it could work and what we could build upon, Johannes came up with the name “Mopidy.” The name is, maybe quite obviously, a combination of the consonants from “MPD” combined with the vowels from “Spotify.” At the same time, the name is different enough from both of its origins not to be mixed up with them. Even during the first few hours we had some thoughts about maybe adding file playback and support for other backends in the future. Thus we quickly appreciated that the “Mopidy” name would still work, even if Spotify wasn’t always the sole focus of the project.
Within a couple of hours we had a Git repo with some plans written up. We joined the #mopidy IRC channel on Freenode and we had recruited Thomas Adamcik to the project. Over the next few years, he designed many of our most essential components, including the extension system. Today, ten years later, Thomas is still involved with Mopidy and many of its extensions.
After a couple of days, it worked! We had built a primitive MPD server in Python that at least worked with the Sonata MPD client. On the backend side, we used the reverse-engineered “despotify” library to interface with Spotify as it already had some Python bindings available. For all three of us, coming mostly from web development and Django, I believe we already had a feeling of achievement and expanding horizons. If we could pull this off, we could build anything.
The story of Mopidy is a story of thousands of small iterative improvements that, over time, add up to something far greater than the sum of its parts. It was a hack, but a hack with good test coverage from the very start, making changes and iteration safe and joyful.
In March 2010 we released our first alpha release. Over the next decade this would become the first of 74 releases, not counting the numerous releases of the extensions we later extracted from Mopidy or built from the ground up.
Later in 2010, as we added an alternative Spotify backend using the official libspotify library, we started seeing the first traces of our current system architecture. At that time, you switched between the single active backend by manually changing the config file and restarting Mopidy. Support for multiple active backends wouldn’t come until much later, after we had built the muxing “core layer” in-between the backend and frontend layers. However, multiple frontends were supported from the start. The MPD server was the first frontend, and during the first year, we added a second one with the Last.fm scrobbler.
By the end of 2010 we had made GStreamer a requirement and thrown out the
support for direct audio output to OSS and ALSA. In 2011 we built upon the
great power of GStreamer to support multiple outputs; this allowed us to play
audio locally whilst also streaming it to a Shoutcast/Icecast server at the same
time. This support for multiple outputs survived for about a year before we
removed it. Instead, we exposed the GStreamer pipeline configuration directly,
just like we still do today with the audio/output configuration. Streaming to
Icecast is also still possible but is involved enough to require specific
documentation for the setup. However, by exposing the GStreamer pipeline
directly, we didn’t have to guess what kind of installations people would use
Mopidy in and exposed the full power of GStreamer to our end-users.
We added support for Python 2.7, but we definitely didn’t plan to stay with it for eight years. During the winter of 2010/2011, I designed and built the Pykka actor library based on the concurrency patterns we had established in Mopidy. When we started using Pykka in Mopidy in March 2011, it already supported Python 3.
Towards the end of the year, we added support for the Ubuntu Sound Menu and the MPRIS D-Bus specification in our third frontend.
Most of 2012 went by without much happening other than a few maintenance releases. However, in November, we released the almost revolutionary Mopidy 0.9 after finally building out the muxing core layer in-between frontends and backends. Depending on the type of request from a frontend, the core layer would either forward the request to the correct backend or, e.g., in the case of search, fan out the request to all backends and then merge the returned search results before passing the result back to the frontend. We had accomplished one of our original goals from the very first day of development: we had a music server that could play music from both Spotify and local files.
Less than a month later, the wheel turned again and we released Mopidy 0.10 with the HTTP frontend. This exposed the full Core API using JSON-RPC over a WebSocket. With this it was suddenly possible to build clients for Mopidy directly, instead of going through MPD.
In a lazy and ingenious moment, we decided that we had no interest in manually keeping the Core API and the JSON-RPC API in sync for the indefinite future. Thus, we based the JSON-RPC API on introspection of the Core API and included a JSON-RPC endpoint which returned a data structure that described the full API. On top of this, I built the Mopidy.js library and released it together with Mopidy 0.10. Mopidy.js uses the API description data structure to dynamically build a mirror of our Core API in JavaScript, working both on Node.js and in the browser.
Even as the JSON-RPC implementation and the Mopidy.js library became the foundation for several popular Mopidy web clients over the next few years, no bug was ever reported that originated in this library. To this day, I testament this to two things: proper test-driven development and excellent code review by Thomas, making these few weeks in November-December 2012 one of the highlights of my years as an open source maintainer.
Jumping just a few months ahead to Easter 2013, the next revolution was about to happen. In a single long and intense day, Thomas and I hashed out and implemented Mopidy’s extension support. Up to this point, there was just one Mopidy. Today, a search for “mopidy” on PyPI returns 127 results.
The Stream backend was created early in 2013, and later in the year, it learned how to parse several playlist formats to find the streamable URL they contained. This made it easy to build backend extensions for music services that exposed playable URIs, like SoundCloud, Google Music, and thousands of radio stations. The backends only had to find and present the playable streams as a playlist or as a virtual file hierarchy; the heavy-lifting of actually playing the audio could be fully delegated to Mopidy and its Stream backend.
By the end of 2013 we had performed the first round of shrinking Mopidy’s core. The Spotify support, Last.fm scrobbler and MPRIS server were all extracted to new extensions living outside the core project. I believe that pulling extensions out of core has helped reduce the amount one must juggle in one’s head to effectively develop on Mopidy.
In 2013 we eased the on-ramping for new users by automatically creating an initial configuration file on the first run. Mopidy also got support for announcing its servers through Zeroconf so they could be autodetected by mobile apps, like Mopidy-Mobile.
Elsewhere in 2013, Wouter van Wijk built the first iterations of the Pi MusicBox distribution for Raspberry Pi. Pi MusicBox provided a turn-key jukebox setup built on Mopidy. This made Mopidy more approachable for the masses that didn’t know Mopidy, Python, or even Linux; allowing them to create their own hi-fi setups.
The next year, in 2014, Fon launched a Kickstarter campaign to build a “modern cloud jukebox” named Gramofon. It turned out that they based their prototype on Mopidy and Javier Domingo Cansino from their development team started submitting patches and becoming active in Mopidy development.
In the summer of 2014 we had our first real-life development sprint at EuroPython in Berlin. Javier and I were joined by several newcomers that got up and running with Mopidy development and squashed a few bugs.
Mopidy 0.19.5 was released on Mopidy’s fifth anniversary in December 2014. A few months later, we released Mopidy 1.0. The release of Mopidy 1.0 did not mark a breaking change, but rather the decision that we could commit to the current APIs for a while and bring stability to the extension ecosystem.
In the summer of 2015, Thomas joined us as we had our second development sprint at EuroPython in Bilbao, Javier’s hometown.
The year between the sprints of 2014 and 2015, and the 0.19, 1.0, and 1.1 releases, were quite significant when looking back at the project’s history. They were not as notable in features as when we added multi-backend, the HTTP API, and extension support back in 2012/2013. Still, they were significant in that the project garnered lots of interest. Up to twenty different people contributed code to each of these three releases.
More than five years ago, in July 2014, I opened Mopidy’s issue #779 for tracking the port to Python 3. There were three large buckets of work that had to be completed. First, our libspotify Python bindings had no Python 3 support. Second, we needed to upgrade to GStreamer 1. Finally, we had to port Mopidy and all of its extensions.
Over the next year, I rewrote pyspotify from scratch using CFFI. Just a couple of weeks before I shipped the final version, Spotify deprecated the libspotify API. However, since they’ve never provided a replacement API for audio playback, we’re still using pyspotify 2 and libspotify to play music from Spotify more than four years later.
During the autumn of 2015 I ported Mopidy from GStreamer 0.10 to 1.x. GStreamer 0.10 was quickly being deprecated, so this work was necessary solely for Mopidy to continue being packaged in Debian and Ubuntu. It was a nice bonus that the new PyGObject wrapper also supported Python 3. With the release of Mopidy 2.0 in February 2016, the move to GStreamer 1 was complete, and thanks to Thomas, we finally supported gapless playback.
Shortly after the 2.0 release, life caught up with several of the most active contributors. A mix of more kids and more work threw Mopidy into a three-year-long period with lower activity and almost exclusively maintenance releases. The only significant development of Mopidy during this period was the support for persisting playback and tracklist state across restarts, contributed by Jens Lütjen, and released in 2.1 in 2017.
Jumping ahead to 2019. Five years after I wrote the tracking issue for moving Mopidy to Python 3, the world looks quite different. Python 2.7’s announced end-of-life is looming at the end of the year, and Python 3 is the standard for all new projects.
So, finally, in the middle of October, we got started on the third and final step toward Python 3.
Once we had a small part of the test suite running on both Python 2 and 3, it took Nick Steel and myself about three weeks of porting modules one by one until suddenly the full test suite was running. Once Mopidy without extensions ran well on Python 3, we axed all Python 2 support, cleaned up all hacks left over from the porting process, and reformatted the code with Black. All of a sudden Mopidy felt like a modern Python codebase.
The six weeks since then have mostly been spent on extensions.
We’ve built a new Mopidy extension registry. We believe that the new registry will ease the discovery of extensions in general. Short-term, we also hope it will help users navigate the extension ecosystem while it is temporarily split in two between Python 2 and 3.
All extensions in the Mopidy GitHub organization are now running on Python 3, as well as a few popular extensions elsewhere. In total, we have almost 20 extensions compatible with Mopidy 3.0 on the day of the release.
Some extensions have also recently received some extra tender loving care.
The Mopidy-Local extension was pushed out of core early in the Python 3 porting. After becoming an independent extension, Thomas Kemmer’s excellent Mopidy-Local-SQLite and Mopidy-Local-Images were merged into Mopidy-Local. We now have a single comprehensive extension for using Mopidy with pre-indexed local music collections.
Next, Nick did a great job fixing up the Mopidy-Spotify extension. Since Spotify suddently broke the playlist part of libspotify a while ago, Mopidy-Spotify has been without functional playlist support. It now uses the Spotify Web API for everything related to playlists and Nick has continued adding several new features using the Web API. Some are shipped in the release that went out together with Mopidy 3.0, and more are right around the corner. Once complete, Mopidy-Spotify will support everything Mopidy-Spotify-Web provides, once again leaving us with a single comprehensive extension for using Mopidy with Spotify.
Just a few days ago, the Mopidy-MPD frontend was pushed out to an extension too. With this, we’ve come full circle from Mopidy being named after “MPD” and “Spotify.” As of Mopidy 3.0, both Mopidy-MPD and Mopidy-Spotify are independent extensions, and Mopidy core knows nothing of either. I wasn’t entirely sure whether moving the MPD server would bring any benefits. Still, once the move was complete, it was evident that Mopidy-MPD deserves to be a project by itself. The split reduced the amount of code in Mopidy by 25% and cut the test suite run time in half. Hopefully this will also make it easier for newcomers to start contributing to either Mopidy-MPD or Mopidy itself.
Finally, December 22, the day before the Mopidy project’s 10th anniversary, we published Mopidy 3.0.
Together with Mopidy 3.0, we uploaded ten updated extensions to PyPI, with at least six more having compatible pre-releases, and, hopefully, final releases over the next few days. For an up to date overview of what’s ready for Python 3 right now, the extension registry is the place to look.
It’s still early days for Mopidy 3.0. Our Homebrew tap is up to date with the new releases, and parts of Arch Linux are already up to date. Updated Debian packages, both at apt.mopidy.com and in Debian, are still some days away. Nonetheless, Mopidy 3.0 will definitively be a part of Ubuntu 20.04 LTS come spring.
Now, go forth and update your extensions to work with Python 3.7+ and Mopidy 3.0.
In a week, Python 2.7 reaches end-of-life. Mopidy, however, is ready for the next decade.
Thanks to Nick Steel for reviewing this blog post.
This blog post was originally published at mopidy.com.
Catalin George Festila: Python 3.7.5 : Django admin shell by Grzegorz Tężycki.
Catalin George Festila: Python 3.7.5 : About Django REST framework.
Python Circle: Using a custom domain for Django app hosted on AWS EC2
Reuven Lerner: Ace Python Interviews — a new, free course to help you get a better job
It’s hard to exaggerate just how hot Python is right now. Lots of companies — from small startups to the Fortune 100 — have realized that Python allows them to do more in less time, and with less code. This means, of course, that companies are scrambling to hire Python developers. There’s tons of demand, and not nearly enough supply.
In other words: Now is a great time to be a Python developer! There are opportunities in just about every field, from Web development to system administration, devops to machine learning, automated testing to financial calculations.
If you’re going to get a Python job, you’ll first have to pass a Python job interview. And like everyone else, you’ll likely prepare for the interview by searching online for “Python interview questions,” or the like.
The good news: There are lots of sites offering Python interview questions and answers.
The bad news: I’ve looked at a lot of them, and they are terrible. The questions are often superficial, and the answers are often wrong or outdated. Plus, a programming interview isn’t a multiple-choice test, in which getting the right answer is the point. Rather, interviewers use the time with you to evaluate your depth of understanding, your coding process, and your ability to adapt as specifications change.
I’ve decided to do my part to change this: Today, I’m launching “Ace Python Interviews,” a new course that covers 50 questions you might be asked on a Python interview. The course has questions for beginner, intermediate, and advanced Python developers. Note that the questions aren’t about specific disciplines, such as Web development or data science; they’re about the core Python language.
The course consists of six hours of video screencasts, written and presented by me while using the Jupyter notebook. And yes, you can download the Jupyter notebooks I used from the course site.
Better yet: This new course is completely free. That’s right: I’m giving this course away, no strings attached. Watch the videos as often (or rarely) as you want — but watch them, learn the process of coding in Python, recognize where you can and should improve your Python skills… and then, go in and knock ’em dead at your interview.
If you’re looking to level up your Python skills, and get a better job using Python in the next year, then I’d suggest taking a look at “Ace Python Interviews.”
Better yet: If you have friends or colleagues who are looking to get a new job with Python, then be sure to mention my new course to them. They might just learn some tips that’ll help them to wow the interviewers, and improve their careers.
Enroll in “Ace Python Interviews”
Join “Ace Python Interviews,” and get a better Python job today!
The post Ace Python Interviews — a new, free course to help you get a better job appeared first on Reuven Lerner.
Podcast.__init__: Python's Built In IDE Isn't Just Sitting IDLE
Summary
One of the first challenges that new programmers are faced with is figuring out what editing environment to use. For the past 20 years, Python has had an easy answer to that question in the form of IDLE. In this episode Tal Einat helps us explore its history, the ways it is used, how it was built, and what is in store for its future. Even if you have never used the IDLE editor yourself, it is still an important piece of Python’s strength and history, and this conversation helps to highlight why that is.
Announcements
- Hello and welcome to Podcast.__init__, the podcast about Python and the people who make it great.
- When you’re ready to launch your next app or want to try a project you hear about on the show, you’ll need somewhere to deploy it, so take a look at our friends over at Linode. With 200 Gbit/s private networking, scalable shared block storage, node balancers, and a 40 Gbit/s public network, all controlled by a brand new API you’ve got everything you need to scale up. And for your tasks that need fast computation, such as training machine learning models, they just launched dedicated CPU instances. Go to pythonpodcast.com/linode to get a $20 credit and launch a new server in under a minute. And don’t forget to thank them for their continued support of this show!
- You listen to this show to learn and stay up to date with the ways that Python is being used, including the latest in machine learning and data analysis. For even more opportunities to meet, listen, and learn from your peers you don’t want to miss out on this year’s conference season. We have partnered with organizations such as O’Reilly Media, Corinium Global Intelligence, ODSC, and Data Council. Upcoming events include the Software Architecture Conference in NYC, Strata Data in San Jose, and PyCon US in Pittsburgh. Go to pythonpodcast.com/conferences to learn more about these and other events, and take advantage of our partner discounts to save money when you register today.
- Your host as usual is Tobias Macey and today I’m interviewing Tal Einat about the IDLE editor for Python, it’s history, and what is in store for its future
Interview
- Introductions
- How did you get introduced to Python?
- For anyone who hasn’t used it, can you start by explaining what IDLE is?
- IDLE has been part of the standard library for Python for quite some time now. What was the motivation for adding it to the core of Python?
- How has the evolution of our computing environment changed the motivation for maintaining IDLE and the use cases that it is most beneficial for?
- What are the benefits of including a basic editor in the default distribution of Python?
- What are some of the ways in which it is often used?
- What are the limiting factors that lead users to other IDEs or text editors?
- What role do you think IDLE has played in the growth of Python?
- What was your motivation for getting involved as a Python contributor and working on the implementation of IDLE?
- How is IDLE implemented and what are some of the ways that it has evolved since its initial introduction?
- How well has the code for IDLE aged as new features and capabilities are added to the language?
- What are some of the integration points available for extending IDLE?
- What are some of the most interesting or innovative ways that you have seen IDLE used and extended?
- What is planned for the future of the IDLE module?
Keep In Touch
Picks
- Tobias
- Tal
- Captain Fantastic
- The Lesson To Unlearn article by Paul Graham
Closing Announcements
- Thank you for listening! Don’t forget to check out our other show, the Data Engineering Podcast for the latest on modern data management.
- Visit the site to subscribe to the show, sign up for the mailing list, and read the show notes.
- If you’ve learned something or tried out a project from the show then tell us about it! Email hosts@podcastinit.com) with your story.
- To help other people find the show please leave a review on iTunes and tell your friends and co-workers
- Join the community in the new Zulip chat workspace at pythonpodcast.com/chat
Links
- IDLE
- FullProof
- Israel
- Eric Idle
- Monty Python
- Visual Studio
- IDLE-fork
- Vi
- Emacs
- Sublime Text
- Visual Studio Code
- REPL == Read Eval Print Loop
- Tcl/Tk
- Tkinter
- RPC == Remote Procedure Call
- IDLEx
- VPython
- Python Turtle
- SVN (Subversion)
The intro and outro music is from Requiem for a Fish The Freak Fandango Orchestra / CC BY-SA
IslandT: Return an even number based on the Nth even number with python
In this example, we will create a python function that will return an even number based on the Nth even number given.
Let say when we enter one into that function, the function will return 0 because the first even number is 0. If we enter two into that function, the function will return 2 because the second number of even numbers is 2. Besides that, we will also need to take care of the number that is smaller than 1 which is an invalid entry as one is the very first even number.
def nth_even(n): if n < 1: return 0 # return the first even number if the user has entered an invalid number else: return 2 * (n-1)
Enter your own solution in the comment box below!
Quansight Labs Blog: metadsl PyData talk
metadsl PyData talk
PyData NYC just ended and I thought it would be good to collect my thoughts on metadsl based on the many conversations I had there surrounding it. This is a rather long post, so if you are just looking for some code here is a Binder link for my talk
What is metadsl?
classNumber(metadsl.Expression):@metadsl.expressiondef__add__(self,other:Number)->Number:...@metadsl.expression@classmethoddeffrom_int(cls,i:int)->Number:...@metadsl.ruledefadd_zero(y:Number):yieldNumber.from_int(0)+y,yyieldy+Number.from_int(0),y
Read more… (12 min remaining to read)
Catalin George Festila: Python 3.7.5 : Is Django the best web framework?
Stack Abuse: How to Get the Current Date and Time in Python
Introduction
Logging, saving records to the database, and accessing files are all common tasks a programmer works on. In each of those cases, date and time play an important role in preserving the meaning and integrity of the data. Programmers often need to engage with date and time.
In this article we will learn how to get the current date and time using Python's builtin datetime module. With that module, we can get all relevant data in one object, or extract the date and time separately.
We will also learn how to adjust our date and time for different timezones. Finally, we'll look at converting datetime objects to the popular Unix or Epoch timestamps.
Getting the Current Date and Time with datetime
The datetime module contains various classes to get information about the date and time:
datetime.date: Stores the day, month and year or a datedatetime.time: Stores the time in hours, minutes, seconds, and microseconds. This information is independent from any datedatetime.datetime: Stores bothdateandtimeattributes
Let's get the current date and time with a datetime.datetime object, as it is easier to extract date and time objects from it. First, let's import the datetime module:
from datetime import datetime
While it looks strange at first, we are getting the datetime class from the datetime module, which are two separate things.
We can use the now() function to get the an object with the current date and time:
current_datetime = datetime.now()
print(current_datetime)
Running this file will give us the following output:
2019-12-13 12:18:18.290623
The now() function gave us an object with all date and time of the moment it was created. When we print it, Python automatically formats the object to a string so it can be read by humans easily. You can read our guide on How to Format Dates in Python to learn various ways to print the time.
You can get individual attributes of the date and time like this:
print(current_datetime.year) # 2019
print(current_datetime.month) # 12
print(current_datetime.day) # 13
print(current_datetime.hour) # 12
print(current_datetime.minute) # 18
print(current_datetime.second) # 18
print(current_datetime.microsecond) # 290623
The now() method is perfect for capturing a moment. However, it is redundant when you only care about either the date or the time, but not both. How can we get date information only?
Getting the Current Date
There are two ways to get the current date in Python. In the Python console, we can get the date from a datetime object with the date() method:
>>> from datetime import datetime
>>> current_date = datetime.now().date()
>>> print(current_date)
2019-12-13
However, it does seem like a waste to initialize a datetime object only to capture the date. An alternative would be to use the today() method of the date class:
>>> from datetime import date
>>> current_date = date.today()
>>> print(current_date)
2019-12-13
The datetime.date class represents a calendar date. Its year, month, and day attributes can be accessed in the same way we access them on the datetime object.
For example, this print statement gets each attribute, and also uses the weekday() method to get the day of the week:
>>> print("It's the %dth day of the week, %dth day of the %dth month of the %dth year" %
... (current_date.weekday()+1, current_date.day, current_date.month, current_date.year))
It's the 5th day of the week, 13th day of the 12th month of the 2019th year
Note: The weekday() method returns an integer between 0 and 6 to represent the day of the week. 0 corresponds to Monday. Therefore, in our example we added 1 in order to print the 5th day of the week (Friday), although the weekday() method returned 4.
Now that we can get the current date by itself, let's see how we can get the current time
Getting the Current Time
We capture the current time from a datetime object using the time() method:
>>> from datetime import datetime
>>> current_time = datetime.now().time()
>>> print(current_time)
12:18:18.290623
Unlike retrieving dates, there is no shortcut to capture current the time directly. The datetime.time class represents a notion of time as in hours, minutes, seconds and microseconds - anything that is less than a day.
Like the datetime object, we can access its attributes directly. Here's an example where we print the current_time in a sentence:
>>> print("It's %d o'clock. To be exact, it's %d:%d and %d seconds with %d microseconds" %
... (current_time.hour, current_time.hour, current_time.minute, current_time.second, current_time.microsecond))
It's 12 o'clock. To be exact, it's 12:18 and 18 seconds with 290623 microseconds
By default, the now() function returns time your local timezone. What if we'd need to convert it to a different one which was set in a user's preference? Let's see how we can provide timezone information to get localized time objects.
Getting the Current Date and Time in Different Timezone
The now() method accepts a timezone as an argument, so that the datetime object being generated would be appropriately adjusted. First, we need to get timezone data via the pytz library.
Install the pytz library with pip if it's not available on your computer already:
$ pip3 install pytz
Now, let's use pytz to get the current date and time if we were in Berlin:
>>> import pytz
>>> from datetime import datetime
>>> tz = pytz.timezone('Europe/Berlin')
>>> berlin_current_datetime = datetime.now(tz)
>>> print(berlin_current_datetime)
2019-12-14 00:20:50.840350+01:00
The berlin_current_datetime would be a datetime object with all the properties and functions we saw earlier, but now it perfectly matches what it is for someone who lives there.
If you would like to get the time in UTC, you can use the pytz module like before:
>>> import pytz
>>> from datetime import datetime
>>> utc_current_datetime = datetime.now(pytz.timezone("UTC"))
>>> print(utc_current_datetime)
2019-12-13 23:20:50.840350+00:00
Alternatively, for UTC time you don't have to use the pytz module at all. The datetime module has a timezone property, which can be used like this:
>>> from datetime import datetime, timezone
>>> utc_current_datetime = datetime.now(timezone.utc)
>>> print(utc_current_datetime)
2019-12-13 23:20:50.840350+00:00
Now that we can convert our dates and times to match different timezones, let's look at how we can convert it to one of the most widely used formats in computing - Unix timestamps.
Converting Timestamps to Dates and Times
Computer systems measure time based on the number of seconds elapsed since the Unix epoch, that is 00:00:00 UTC on January 1st, 1970. It's common for applications, databases, and protocols to use a timestamp in order to display time.
You can get the current timestamp in Python using the time library:
>>> import time
>>> print(time.time())
1576280665.0915806
The time.time() function returns a float number with the Unix timestamp. You can convert a timestamp to a datetime object using the fromtimestamp() function:
>>> from datetime import datetime
>>> print(datetime.fromtimestamp(1576280665))
2019-12-13 19:44:25
By default, the fromtimestamp() returns a datetime object in your timezone. You can provide a timezone as a second argument if you would like to have it localized differently.
For example, to convert a timestamp to a datetime object in UTC, you can do this:
>>> from datetime import datetime, timezone
>>> print(datetime.fromtimestamp(1576280665, timezone.utc))
2019-12-13 19:44:25+00:00
Conversely, you can use the timestamp() method to convert a datetime object to a valid timestamp:
>>> from datetime import datetime, timezone
>>> print(datetime.now(timezone.utc).timestamp())
1577117555.940705
Conclusion
Pythons built-in datetime library allows us to get the current date and time with the now() function. We can use the returned datetime object to extract just the date portion or the time portion. We can also get the current date with the today() function.
By default, Python creates datetime objects in our local timezone. If we utilize the pytz library, we can convert dates and times into different timezones.
Finally, we used the fromtimestamp() function to take a Unix timestamp to get a datetime object. If we have a datetime object, we can get a timestamp using its timestamp() method.
Real Python: Python Dictionary Iteration: Advanced Tips & Tricks
Dictionaries are one of the most important and useful data structures in Python. They can help you solve a wide variety of programming problems. This course will take you on a deep dive into how to iterate through a dictionary in Python.
By the end of this course, you’ll know:
- What dictionaries are, as well as some of their main features and implementation details
- How to iterate through a dictionary in Python by using the basic tools the language offers
- What kind of real-world tasks you can perform by iterating through a dictionary in Python
- How to use some more advanced techniques and strategies to iterate through a dictionary in Python
For more information on dictionaries, you can check out the following resources:
- Dictionaries in Python
- Itertools in Python 3, By Example
- The documentation for
map()andfilter()
Ready? Let’s go!
[ Improve Your Python With 🐍 Python Tricks 💌 – Get a short & sweet Python Trick delivered to your inbox every couple of days. >> Click here to learn more and see examples ]
PyPy Development: PyPy 7.3.0 released
- PyPy2.7, which is an interpreter supporting the syntax and the features of Python 2.7 including the stdlib for CPython 2.7.13
- PyPy3.6: which is an interpreter supporting the syntax and the features of Python 3.6, including the stdlib for CPython 3.6.9.
The interpreters are based on much the same codebase, thus the double release.
We have worked with the python packaging group to support tooling around building third party packages for python, so this release changes the ABI tag for PyPy.
Based on the great work done in portable-pypy, the linux downloads we provide are now built on top of the manylinux2010 CentOS6 docker image. The tarballs include the needed shared objects to run on any platform that supports manylinux2010 wheels, which should include all supported versions of debian- and RedHat-based distributions (including Ubuntu, CentOS, and Fedora).
The CFFI backend has been updated to version 1.13.1. We recommend using CFFI rather than c-extensions to interact with C.
The built-in
cppyy module was upgraded to 1.10.6, which
provides, among others, better template resolution, stricter enum handling,
anonymous struct/unions, cmake fragments for distribution, optimizations for
PODs, and faster wrapper calls. We reccomend using cppyy for performant
wrapping of C++ code for Python.The vendored pyrepl package for interaction inside the REPL was updated.
Support for codepage encoding and decoding was added for Windows.
As always, this release fixed several issues and bugs raised by the growing community of PyPy users. We strongly recommend updating. Many of the fixes are the direct result of end-user bug reports, so please continue reporting issues as they crop up.
You can download the v7.3 releases here:
We would like to thank our donors for the continued support of the PyPy project. If PyPy is not quite good enough for your needs, we are available for direct consulting work.
We would also like to thank our contributors and encourage new people to join the project. PyPy has many layers and we need help with all of them: PyPy and RPython documentation improvements, tweaking popular packages to run on pypy, or general help with making RPython’s JIT even better. Since the previous release, we have accepted contributions from 3 new contributors, thanks for pitching in.
If you are a python library maintainer and use c-extensions, please consider making a cffi / cppyy version of your library that would be performant on PyPy. If you are stuck with using the C-API, you can use docker images with PyPy built in or the multibuild system to build wheels.
What is PyPy?
PyPy is a very compliant Python interpreter, almost a drop-in replacement for CPython 2.7, 3.6. It’s fast (PyPy and CPython 2.7.x performance comparison) due to its integrated tracing JIT compiler.We also welcome developers of other dynamic languages to see what RPython can do for them.
This PyPy release supports:
- x86 machines on most common operating systems (Linux 32/64 bit, Mac OS X 64-bit, Windows 32-bit, OpenBSD, FreeBSD)
- big- and little-endian variants of PPC64 running Linux
- s390x running Linux
- 64-bit ARM machines running Linux
What else is new?
PyPy 7.2 was released in October, 2019. There are many incremental improvements to RPython and PyPy, For more information about the 7.3.0 release, see the full changelog.Please update, and continue to help us make PyPy better.
Cheers,
The PyPy team









