Pandas is a powerful and flexible Python package that allows you to work with labeled and time series data. It also provides statistics methods, enables plotting, and more. One crucial feature of Pandas is its ability to write and read Excel, CSV, and many other types of files. Functions like the Pandas read_csv() method enable you to work with files effectively. You can use them to save the data and labels from Pandas objects to a file and load them later as Pandas Series or DataFrame instances.
In this tutorial, you’ll learn:
- What the Pandas IO tools API is
- How to read and write data to and from files
- How to work with various file formats
- How to work with big data efficiently
Let’s start reading and writing files!
Free Bonus:5 Thoughts On Python Mastery, a free course for Python developers that shows you the roadmap and the mindset you'll need to take your Python skills to the next level.
Installing Pandas
The code in this tutorial is executed with CPython 3.7.4 and Pandas 0.25.1. It would be beneficial to make sure you have the latest versions of Python and Pandas on your machine. You might want to create a new virtual environment and install the dependencies for this tutorial.
First, you’ll need the Pandas library. You may already have it installed. If you don’t, then you can install it with pip:
Once the installation process completes, you should have Pandas installed and ready.
Anaconda is an excellent Python distribution that comes with Python, many useful packages like Pandas, and a package and environment manager called Conda. To learn more about Anaconda, check out Setting Up Python for Machine Learning on Windows.
If you don’t have Pandas in your virtual environment, then you can install it with Conda:
Conda is powerful as it manages the dependencies and their versions. To learn more about working with Conda, you can check out the official documentation.
Preparing Data
In this tutorial, you’ll use the data related to 20 countries. Here’s an overview of the data and sources you’ll be working with:
Country is denoted by the country name. Each country is in the top 10 list for either population, area, or gross domestic product (GDP). The row labels for the dataset are the three-letter country codes defined in ISO 3166-1. The column label for the dataset is COUNTRY.
Population is expressed in millions. The data comes from a list of countries and dependencies by population on Wikipedia. The column label for the dataset is POP.
Area is expressed in thousands of kilometers squared. The data comes from a list of countries and dependencies by area on Wikipedia. The column label for the dataset is AREA.
Gross domestic product is expressed in millions of U.S. dollars, according to the United Nations data for 2017. You can find this data in the list of countries by nominal GDP on Wikipedia. The column label for the dataset is GDP.
Continent is either Africa, Asia, Oceania, Europe, North America, or South America. You can find this information on Wikipedia as well. The column label for the dataset is CONT.
Independence day is a date that commemorates a nation’s independence. The data comes from the list of national independence days on Wikipedia. The dates are shown in ISO 8601 format. The first four digits represent the year, the next two numbers are the month, and the last two are for the day of the month. The column label for the dataset is IND_DAY.
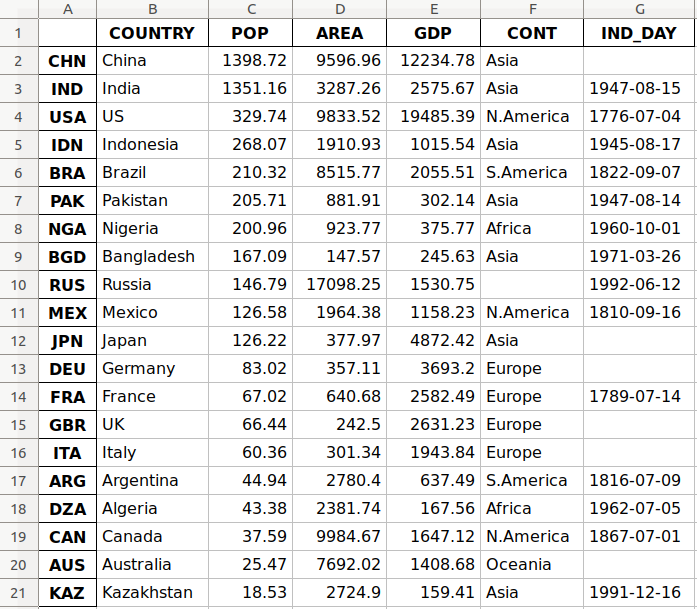
This is how the data looks as a table:
| COUNTRY | POP | AREA | GDP | CONT | IND_DAY |
|---|
| CHN | China | 1398.72 | 9596.96 | 12234.78 | Asia | |
| IND | India | 1351.16 | 3287.26 | 2575.67 | Asia | 1947-08-15 |
| USA | US | 329.74 | 9833.52 | 19485.39 | N.America | 1776-07-04 |
| IDN | Indonesia | 268.07 | 1910.93 | 1015.54 | Asia | 1945-08-17 |
| BRA | Brazil | 210.32 | 8515.77 | 2055.51 | S.America | 1822-09-07 |
| PAK | Pakistan | 205.71 | 881.91 | 302.14 | Asia | 1947-08-14 |
| NGA | Nigeria | 200.96 | 923.77 | 375.77 | Africa | 1960-10-01 |
| BGD | Bangladesh | 167.09 | 147.57 | 245.63 | Asia | 1971-03-26 |
| RUS | Russia | 146.79 | 17098.25 | 1530.75 | | 1992-06-12 |
| MEX | Mexico | 126.58 | 1964.38 | 1158.23 | N.America | 1810-09-16 |
| JPN | Japan | 126.22 | 377.97 | 4872.42 | Asia | |
| DEU | Germany | 83.02 | 357.11 | 3693.20 | Europe | |
| FRA | France | 67.02 | 640.68 | 2582.49 | Europe | 1789-07-14 |
| GBR | UK | 66.44 | 242.50 | 2631.23 | Europe | |
| ITA | Italy | 60.36 | 301.34 | 1943.84 | Europe | |
| ARG | Argentina | 44.94 | 2780.40 | 637.49 | S.America | 1816-07-09 |
| DZA | Algeria | 43.38 | 2381.74 | 167.56 | Africa | 1962-07-05 |
| CAN | Canada | 37.59 | 9984.67 | 1647.12 | N.America | 1867-07-01 |
| AUS | Australia | 25.47 | 7692.02 | 1408.68 | Oceania | |
| KAZ | Kazakhstan | 18.53 | 2724.90 | 159.41 | Asia | 1991-12-16 |
You may notice that some of the data is missing. For example, the continent for Russia is not specified because it spreads across both Europe and Asia. There are also several missing independence days because the data source omits them.
You can organize this data in Python using a nested dictionary:
data={'CHN':{'COUNTRY':'China','POP':1_398.72,'AREA':9_596.96,'GDP':12_234.78,'CONT':'Asia'},'IND':{'COUNTRY':'India','POP':1_351.16,'AREA':3_287.26,'GDP':2_575.67,'CONT':'Asia','IND_DAY':'1947-08-15'},'USA':{'COUNTRY':'US','POP':329.74,'AREA':9_833.52,'GDP':19_485.39,'CONT':'N.America','IND_DAY':'1776-07-04'},'IDN':{'COUNTRY':'Indonesia','POP':268.07,'AREA':1_910.93,'GDP':1_015.54,'CONT':'Asia','IND_DAY':'1945-08-17'},'BRA':{'COUNTRY':'Brazil','POP':210.32,'AREA':8_515.77,'GDP':2_055.51,'CONT':'S.America','IND_DAY':'1822-09-07'},'PAK':{'COUNTRY':'Pakistan','POP':205.71,'AREA':881.91,'GDP':302.14,'CONT':'Asia','IND_DAY':'1947-08-14'},'NGA':{'COUNTRY':'Nigeria','POP':200.96,'AREA':923.77,'GDP':375.77,'CONT':'Africa','IND_DAY':'1960-10-01'},'BGD':{'COUNTRY':'Bangladesh','POP':167.09,'AREA':147.57,'GDP':245.63,'CONT':'Asia','IND_DAY':'1971-03-26'},'RUS':{'COUNTRY':'Russia','POP':146.79,'AREA':17_098.25,'GDP':1_530.75,'IND_DAY':'1992-06-12'},'MEX':{'COUNTRY':'Mexico','POP':126.58,'AREA':1_964.38,'GDP':1_158.23,'CONT':'N.America','IND_DAY':'1810-09-16'},'JPN':{'COUNTRY':'Japan','POP':126.22,'AREA':377.97,'GDP':4_872.42,'CONT':'Asia'},'DEU':{'COUNTRY':'Germany','POP':83.02,'AREA':357.11,'GDP':3_693.20,'CONT':'Europe'},'FRA':{'COUNTRY':'France','POP':67.02,'AREA':640.68,'GDP':2_582.49,'CONT':'Europe','IND_DAY':'1789-07-14'},'GBR':{'COUNTRY':'UK','POP':66.44,'AREA':242.50,'GDP':2_631.23,'CONT':'Europe'},'ITA':{'COUNTRY':'Italy','POP':60.36,'AREA':301.34,'GDP':1_943.84,'CONT':'Europe'},'ARG':{'COUNTRY':'Argentina','POP':44.94,'AREA':2_780.40,'GDP':637.49,'CONT':'S.America','IND_DAY':'1816-07-09'},'DZA':{'COUNTRY':'Algeria','POP':43.38,'AREA':2_381.74,'GDP':167.56,'CONT':'Africa','IND_DAY':'1962-07-05'},'CAN':{'COUNTRY':'Canada','POP':37.59,'AREA':9_984.67,'GDP':1_647.12,'CONT':'N.America','IND_DAY':'1867-07-01'},'AUS':{'COUNTRY':'Australia','POP':25.47,'AREA':7_692.02,'GDP':1_408.68,'CONT':'Oceania'},'KAZ':{'COUNTRY':'Kazakhstan','POP':18.53,'AREA':2_724.90,'GDP':159.41,'CONT':'Asia','IND_DAY':'1991-12-16'}}columns=('COUNTRY','POP','AREA','GDP','CONT','IND_DAY')Each row of the table is written as an inner dictionary whose keys are the column names and values are the corresponding data. These dictionaries are then collected as the values in the outer data dictionary. The corresponding keys for data are the three-letter country codes.
You can use this data to create an instance of a Pandas DataFrame. First, you need to import Pandas:
Now that you have Pandas imported, you can use the DataFrame constructor and data to create a DataFrame object.
data is organized in such a way that the country codes correspond to columns. You can reverse the rows and columns of a DataFrame with the property .T:
>>>>>> df=pd.DataFrame(data=data).T>>> df COUNTRY POP AREA GDP CONT IND_DAYCHN China 1398.72 9596.96 12234.8 Asia NaNIND India 1351.16 3287.26 2575.67 Asia 1947-08-15USA US 329.74 9833.52 19485.4 N.America 1776-07-04IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26RUS Russia 146.79 17098.2 1530.75 NaN 1992-06-12MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16JPN Japan 126.22 377.97 4872.42 Asia NaNDEU Germany 83.02 357.11 3693.2 Europe NaNFRA France 67.02 640.68 2582.49 Europe 1789-07-14GBR UK 66.44 242.5 2631.23 Europe NaNITA Italy 60.36 301.34 1943.84 Europe NaNARG Argentina 44.94 2780.4 637.49 S.America 1816-07-09DZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01AUS Australia 25.47 7692.02 1408.68 Oceania NaNKAZ Kazakhstan 18.53 2724.9 159.41 Asia 1991-12-16
Now you have your DataFrame object populated with the data about each country.
Note: You can use .transpose() instead of .T to reverse the rows and columns of your dataset. If you use .transpose(), then you can set the optional parameter copy to specify if you want to copy the underlying data. The default behavior is False.
Versions of Python older than 3.6 did not guarantee the order of keys in dictionaries. To ensure the order of columns is maintained for older versions of Python and Pandas, you can specify index=columns:
>>>>>> df=pd.DataFrame(data=data,index=columns).T
Now that you’ve prepared your data, you’re ready to start working with files!
Using the Pandas read_csv() and .to_csv() Functions
A comma-separated values (CSV) file is a plaintext file with a .csv extension that holds tabular data. This is one of the most popular file formats for storing large amounts of data. Each row of the CSV file represents a single table row. The values in the same row are by default separated with commas, but you could change the separator to a semicolon, tab, space, or some other character.
Write a CSV File
You can save your Pandas DataFrame as a CSV file with .to_csv():
>>>>>> df.to_csv('data.csv') That’s it! You’ve created the file data.csv in your current working directory. You can expand the code block below to see how your CSV file should look:
,COUNTRY,POP,AREA,GDP,CONT,IND_DAYCHN,China,1398.72,9596.96,12234.78,Asia,IND,India,1351.16,3287.26,2575.67,Asia,1947-08-15USA,US,329.74,9833.52,19485.39,N.America,1776-07-04IDN,Indonesia,268.07,1910.93,1015.54,Asia,1945-08-17BRA,Brazil,210.32,8515.77,2055.51,S.America,1822-09-07PAK,Pakistan,205.71,881.91,302.14,Asia,1947-08-14NGA,Nigeria,200.96,923.77,375.77,Africa,1960-10-01BGD,Bangladesh,167.09,147.57,245.63,Asia,1971-03-26RUS,Russia,146.79,17098.25,1530.75,,1992-06-12MEX,Mexico,126.58,1964.38,1158.23,N.America,1810-09-16JPN,Japan,126.22,377.97,4872.42,Asia,DEU,Germany,83.02,357.11,3693.2,Europe,FRA,France,67.02,640.68,2582.49,Europe,1789-07-14GBR,UK,66.44,242.5,2631.23,Europe,ITA,Italy,60.36,301.34,1943.84,Europe,ARG,Argentina,44.94,2780.4,637.49,S.America,1816-07-09DZA,Algeria,43.38,2381.74,167.56,Africa,1962-07-05CAN,Canada,37.59,9984.67,1647.12,N.America,1867-07-01AUS,Australia,25.47,7692.02,1408.68,Oceania,KAZ,Kazakhstan,18.53,2724.9,159.41,Asia,1991-12-16
This text file contains the data separated with commas. The first column contains the row labels. In some cases, you’ll find them irrelevant. If you don’t want to keep them, then you can pass the argument index=False to .to_csv().
Read a CSV File
Once your data is saved in a CSV file, you’ll likely want to load and use it from time to time. You can do that with the Pandas read_csv() function:
>>>>>> df=pd.read_csv('data.csv',index_col=0)>>> df COUNTRY POP AREA GDP CONT IND_DAYCHN China 1398.72 9596.96 12234.78 Asia NaNIND India 1351.16 3287.26 2575.67 Asia 1947-08-15USA US 329.74 9833.52 19485.39 N.America 1776-07-04IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26RUS Russia 146.79 17098.25 1530.75 NaN 1992-06-12MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16JPN Japan 126.22 377.97 4872.42 Asia NaNDEU Germany 83.02 357.11 3693.20 Europe NaNFRA France 67.02 640.68 2582.49 Europe 1789-07-14GBR UK 66.44 242.50 2631.23 Europe NaNITA Italy 60.36 301.34 1943.84 Europe NaNARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09DZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01AUS Australia 25.47 7692.02 1408.68 Oceania NaNKAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16 In this case, the Pandas read_csv() function returns a new DataFrame with the data and labels from the file data.csv, which you specified with the first argument. This string can be any valid path, including URLs.
The parameter index_col specifies the column from the CSV file that contains the row labels. You assign a zero-based column index to this parameter. You should determine the value of index_col when the CSV file contains the row labels to avoid loading them as data.
You’ll learn more about using Pandas with CSV files later on in this tutorial. You can also check out Reading and Writing CSV Files in Python to see how to handle CSV files with the built-in Python library csv as well.
Using Pandas to Write and Read Excel Files
Microsoft Excel is probably the most widely-used spreadsheet software. While older versions used binary .xls files, Excel 2007 introduced the new XML-based .xlsx file. You can read and write Excel files in Pandas, similar to CSV files. However, you’ll need to install the following Python packages first:
You can install them using pip with a single command:
$ pip install xlwt openpyxl xlsxwriter xlrd
You can also use Conda:
$ conda install xlwt openpyxl xlsxwriter xlrd
Please note that you don’t have to install all these packages. For example, you don’t need both openpyxl and XlsxWriter. If you’re going to work just with .xls files, then you don’t need any of them! However, if you intend to work only with .xlsx files, then you’re going to need at least one of them, but not xlwt. Take some time to decide which packages are right for your project.
Write an Excel File
Once you have those packages installed, you can save your DataFrame in an Excel file with .to_excel():
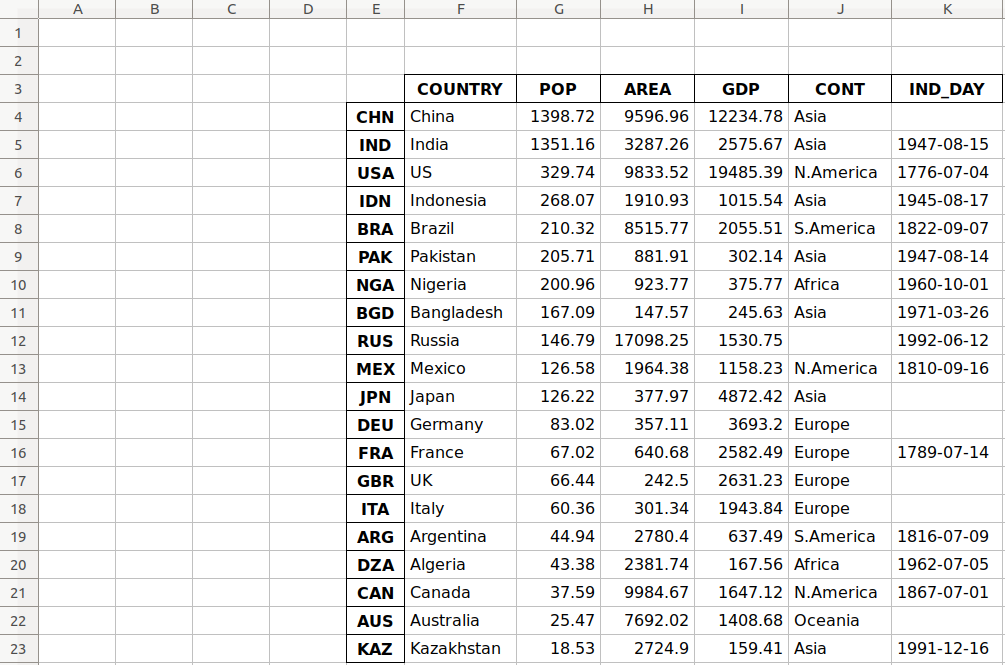
>>>>>> df.to_excel('data.xlsx') The argument 'data.xlsx' represents the target file and, optionally, its path. The above statement should create the file data.xlsx in your current working directory. That file should look like this:
![mmst-pandas-rw-files-excel]()
The first column of the file contains the labels of the rows, while the other columns store data.
Read an Excel File
You can load data from Excel files with read_excel():
>>>>>> df=pd.read_excel('data.xlsx',index_col=0)>>> df COUNTRY POP AREA GDP CONT IND_DAYCHN China 1398.72 9596.96 12234.78 Asia NaNIND India 1351.16 3287.26 2575.67 Asia 1947-08-15USA US 329.74 9833.52 19485.39 N.America 1776-07-04IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26RUS Russia 146.79 17098.25 1530.75 NaN 1992-06-12MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16JPN Japan 126.22 377.97 4872.42 Asia NaNDEU Germany 83.02 357.11 3693.20 Europe NaNFRA France 67.02 640.68 2582.49 Europe 1789-07-14GBR UK 66.44 242.50 2631.23 Europe NaNITA Italy 60.36 301.34 1943.84 Europe NaNARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09DZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01AUS Australia 25.47 7692.02 1408.68 Oceania NaNKAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16 read_excel() returns a new DataFrame that contains the values from data.xlsx. You can also use read_excel() with OpenDocument spreadsheets, or .ods files.
You’ll learn more about working with Excel files later on in this tutorial. You can also check out Using Pandas to Read Large Excel Files in Python.
Understanding the Pandas IO API
Pandas IO Tools is the API that allows you to save the contents of Series and DataFrame objects to the clipboard, objects, or files of various types. It also enables loading data from the clipboard, objects, or files.
Write Files
Series and DataFrame objects have methods that enable writing data and labels to the clipboard or files. They’re named with the pattern .to_<file-type>(), where <file-type> is the type of the target file.
You’ve learned about .to_csv() and .to_excel(), but there are others, including:
.to_json().to_html().to_sql().to_pickle()
There are still more file types that you can write to, so this list is not exhaustive.
Note: To find similar methods, check the official documentation about serialization, IO, and conversion related to Series and DataFrame objects.
These methods have parameters specifying the target file path where you saved the data and labels. This is mandatory in some cases and optional in others. If this option is available and you choose to omit it, then the methods return the objects (like strings or iterables) with the contents of DataFrame instances.
The optional parameter compression decides how to compress the file with the data and labels. You’ll learn more about it later on. There are a few other parameters, but they’re mostly specific to one or several methods. You won’t go into them in detail here.
Read Files
Pandas functions for reading the contents of files are named using the pattern .read_<file-type>(), where <file-type> indicates the type of the file to read. You’ve already seen the Pandas read_csv() and read_excel() functions. Here are a few others:
read_json()read_html()read_sql()read_pickle()
These functions have a parameter that specifies the target file path. It can be any valid string that represents the path, either on a local machine or in a URL. Other objects are also acceptable depending on the file type.
The optional parameter compression determines the type of decompression to use for the compressed files. You’ll learn about it later on in this tutorial. There are other parameters, but they’re specific to one or several functions. You won’t go into them in detail here.
Working With Different File Types
The Pandas library offers a wide range of possibilities for saving your data to files and loading data from files. In this section, you’ll learn more about working with CSV and Excel files. You’ll also see how to use other types of files, like JSON, web pages, databases, and Python pickle files.
CSV Files
You’ve already learned how to read and write CSV files. Now let’s dig a little deeper into the details. When you use .to_csv() to save your DataFrame, you can provide an argument for the parameter path_or_buff to specify the path, name, and extension of the target file.
path_or_buff is the first argument .to_csv() will get. It can be any string that represents a valid file path that includes the file name and its extension. You’ve seen this in a previous example. However, if you omit path_or_buff, then .to_csv() won’t create any files. Instead, it’ll return the corresponding string:
>>>>>> df=pd.DataFrame(data=data).T>>> s=df.to_csv()>>> print(s),COUNTRY,POP,AREA,GDP,CONT,IND_DAYCHN,China,1398.72,9596.96,12234.78,Asia,IND,India,1351.16,3287.26,2575.67,Asia,1947-08-15USA,US,329.74,9833.52,19485.39,N.America,1776-07-04IDN,Indonesia,268.07,1910.93,1015.54,Asia,1945-08-17BRA,Brazil,210.32,8515.77,2055.51,S.America,1822-09-07PAK,Pakistan,205.71,881.91,302.14,Asia,1947-08-14NGA,Nigeria,200.96,923.77,375.77,Africa,1960-10-01BGD,Bangladesh,167.09,147.57,245.63,Asia,1971-03-26RUS,Russia,146.79,17098.25,1530.75,,1992-06-12MEX,Mexico,126.58,1964.38,1158.23,N.America,1810-09-16JPN,Japan,126.22,377.97,4872.42,Asia,DEU,Germany,83.02,357.11,3693.2,Europe,FRA,France,67.02,640.68,2582.49,Europe,1789-07-14GBR,UK,66.44,242.5,2631.23,Europe,ITA,Italy,60.36,301.34,1943.84,Europe,ARG,Argentina,44.94,2780.4,637.49,S.America,1816-07-09DZA,Algeria,43.38,2381.74,167.56,Africa,1962-07-05CAN,Canada,37.59,9984.67,1647.12,N.America,1867-07-01AUS,Australia,25.47,7692.02,1408.68,Oceania,KAZ,Kazakhstan,18.53,2724.9,159.41,Asia,1991-12-16
Now you have the string s instead of a CSV file. You also have some missing values in your DataFrame object. For example, the continent for Russia and the independence days for several countries (China, Japan, and so on) are not available. In data science and machine learning, you must handle missing values carefully. Pandas excels here! By default, Pandas uses the NaN value to replace the missing values.
Note:nan, which stands for “not a number,” is a particular floating-point value in Python.
You can get a nan value with any of the following functions:
The continent that corresponds to Russia in df is nan:
>>>>>> df.loc['RUS','CONT']nan
This example uses .loc[] to get data with the specified row and column names.
When you save your DataFrame to a CSV file, empty strings ('') will represent the missing data. You can see this both in your file data.csv and in the string s. If you want to change this behavior, then use the optional parameter na_rep:
>>>>>> df.to_csv('new-data.csv',na_rep='(missing)') This code produces the file new-data.csv where the missing values are no longer empty strings. You can expand the code block below to see how this file should look:
,COUNTRY,POP,AREA,GDP,CONT,IND_DAYCHN,China,1398.72,9596.96,12234.78,Asia,(missing)IND,India,1351.16,3287.26,2575.67,Asia,1947-08-15USA,US,329.74,9833.52,19485.39,N.America,1776-07-04IDN,Indonesia,268.07,1910.93,1015.54,Asia,1945-08-17BRA,Brazil,210.32,8515.77,2055.51,S.America,1822-09-07PAK,Pakistan,205.71,881.91,302.14,Asia,1947-08-14NGA,Nigeria,200.96,923.77,375.77,Africa,1960-10-01BGD,Bangladesh,167.09,147.57,245.63,Asia,1971-03-26RUS,Russia,146.79,17098.25,1530.75,(missing),1992-06-12MEX,Mexico,126.58,1964.38,1158.23,N.America,1810-09-16JPN,Japan,126.22,377.97,4872.42,Asia,(missing)DEU,Germany,83.02,357.11,3693.2,Europe,(missing)FRA,France,67.02,640.68,2582.49,Europe,1789-07-14GBR,UK,66.44,242.5,2631.23,Europe,(missing)ITA,Italy,60.36,301.34,1943.84,Europe,(missing)ARG,Argentina,44.94,2780.4,637.49,S.America,1816-07-09DZA,Algeria,43.38,2381.74,167.56,Africa,1962-07-05CAN,Canada,37.59,9984.67,1647.12,N.America,1867-07-01AUS,Australia,25.47,7692.02,1408.68,Oceania,(missing)KAZ,Kazakhstan,18.53,2724.9,159.41,Asia,1991-12-16
Now, the string '(missing)' in the file corresponds to the nan values from df.
When Pandas reads files, it considers the empty string ('') and a few others as missing values by default:
'nan''-nan''NA''N/A''NaN''null'
If you don’t want this behavior, then you can pass keep_default_na=False to the Pandas read_csv() function. To specify other labels for missing values, use the parameter na_values:
>>>>>> pd.read_csv('new-data.csv',index_col=0,na_values='(missing)') COUNTRY POP AREA GDP CONT IND_DAYCHN China 1398.72 9596.96 12234.78 Asia NaNIND India 1351.16 3287.26 2575.67 Asia 1947-08-15USA US 329.74 9833.52 19485.39 N.America 1776-07-04IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26RUS Russia 146.79 17098.25 1530.75 NaN 1992-06-12MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16JPN Japan 126.22 377.97 4872.42 Asia NaNDEU Germany 83.02 357.11 3693.20 Europe NaNFRA France 67.02 640.68 2582.49 Europe 1789-07-14GBR UK 66.44 242.50 2631.23 Europe NaNITA Italy 60.36 301.34 1943.84 Europe NaNARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09DZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01AUS Australia 25.47 7692.02 1408.68 Oceania NaNKAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16 Here, you’ve marked the string '(missing)' as a new missing data label, and Pandas replaced it with nan when it read the file.
When you load data from a file, Pandas assigns the data types to the values of each column by default. You can check these types with .dtypes:
>>>>>> df=pd.read_csv('data.csv',index_col=0)>>> df.dtypesCOUNTRY objectPOP float64AREA float64GDP float64CONT objectIND_DAY objectdtype: object The columns with strings and dates ('COUNTRY', 'CONT', and 'IND_DAY') have the data type object. Meanwhile, the numeric columns contain 64-bit floating-point numbers (float64).
You can use the parameter dtype to specify the desired data types and parse_dates to force use of datetimes:
>>>>>> dtypes={'POP':'float32','AREA':'float32','GDP':'float32'}>>> df=pd.read_csv('data.csv',index_col=0,dtype=dtypes,... parse_dates=['IND_DAY'])>>> df.dtypesCOUNTRY objectPOP float32AREA float32GDP float32CONT objectIND_DAY datetime64[ns]dtype: object>>> df['IND_DAY']CHN NaTIND 1947-08-15USA 1776-07-04IDN 1945-08-17BRA 1822-09-07PAK 1947-08-14NGA 1960-10-01BGD 1971-03-26RUS 1992-06-12MEX 1810-09-16JPN NaTDEU NaTFRA 1789-07-14GBR NaTITA NaTARG 1816-07-09DZA 1962-07-05CAN 1867-07-01AUS NaTKAZ 1991-12-16Name: IND_DAY, dtype: datetime64[ns] Now, you have 32-bit floating-point numbers ()float32) as specified with dtype. These differ slightly from the original 64-bit numbers because of smaller precision. The values in the last column are considered as dates and have the data type datetime64. That’s why the NaN values in this column are replaced with NaT.
Now that you have real dates, you can save them in the format you like:
>>>>>> df=pd.read_csv('data.csv',index_col=0,parse_dates=['IND_DAY'])>>> df.to_csv('formatted-data.csv',date_format='%B %d, %Y') Here, you’ve specified the parameter date_format to be '%B %d, %Y'. You can expand the code block below to see the resulting file:
,COUNTRY,POP,AREA,GDP,CONT,IND_DAYCHN,China,1398.72,9596.96,12234.78,Asia,IND,India,1351.16,3287.26,2575.67,Asia,"August 15, 1947"USA,US,329.74,9833.52,19485.39,N.America,"July 04, 1776"IDN,Indonesia,268.07,1910.93,1015.54,Asia,"August 17, 1945"BRA,Brazil,210.32,8515.77,2055.51,S.America,"September 07, 1822"PAK,Pakistan,205.71,881.91,302.14,Asia,"August 14, 1947"NGA,Nigeria,200.96,923.77,375.77,Africa,"October 01, 1960"BGD,Bangladesh,167.09,147.57,245.63,Asia,"March 26, 1971"RUS,Russia,146.79,17098.25,1530.75,,"June 12, 1992"MEX,Mexico,126.58,1964.38,1158.23,N.America,"September 16, 1810"JPN,Japan,126.22,377.97,4872.42,Asia,DEU,Germany,83.02,357.11,3693.2,Europe,FRA,France,67.02,640.68,2582.49,Europe,"July 14, 1789"GBR,UK,66.44,242.5,2631.23,Europe,ITA,Italy,60.36,301.34,1943.84,Europe,ARG,Argentina,44.94,2780.4,637.49,S.America,"July 09, 1816"DZA,Algeria,43.38,2381.74,167.56,Africa,"July 05, 1962"CAN,Canada,37.59,9984.67,1647.12,N.America,"July 01, 1867"AUS,Australia,25.47,7692.02,1408.68,Oceania,KAZ,Kazakhstan,18.53,2724.9,159.41,Asia,"December 16, 1991"
The format of the dates is different now. The format '%B %d, %Y' means the date will first display the full name of the month, then the day followed by a comma, and finally the full year.
There are several other optional parameters that you can use with .to_csv():
sep denotes a values separator.decimal indicates a decimal separator.encoding sets the file encoding.header specifies whether you want to write column labels in the file.
Here’s how you would pass arguments for sep and header:
>>>>>> s=df.to_csv(sep=';',header=False)>>> print(s)CHN;China;1398.72;9596.96;12234.78;Asia;IND;India;1351.16;3287.26;2575.67;Asia;1947-08-15USA;US;329.74;9833.52;19485.39;N.America;1776-07-04IDN;Indonesia;268.07;1910.93;1015.54;Asia;1945-08-17BRA;Brazil;210.32;8515.77;2055.51;S.America;1822-09-07PAK;Pakistan;205.71;881.91;302.14;Asia;1947-08-14NGA;Nigeria;200.96;923.77;375.77;Africa;1960-10-01BGD;Bangladesh;167.09;147.57;245.63;Asia;1971-03-26RUS;Russia;146.79;17098.25;1530.75;;1992-06-12MEX;Mexico;126.58;1964.38;1158.23;N.America;1810-09-16JPN;Japan;126.22;377.97;4872.42;Asia;DEU;Germany;83.02;357.11;3693.2;Europe;FRA;France;67.02;640.68;2582.49;Europe;1789-07-14GBR;UK;66.44;242.5;2631.23;Europe;ITA;Italy;60.36;301.34;1943.84;Europe;ARG;Argentina;44.94;2780.4;637.49;S.America;1816-07-09DZA;Algeria;43.38;2381.74;167.56;Africa;1962-07-05CAN;Canada;37.59;9984.67;1647.12;N.America;1867-07-01AUS;Australia;25.47;7692.02;1408.68;Oceania;KAZ;Kazakhstan;18.53;2724.9;159.41;Asia;1991-12-16
The data is separated with a semicolon (';') because you’ve specified sep=';'. Also, since you passed header=False, you see your data without the header row of column names.
The Pandas read_csv() function has many additional options for managing missing data, working with dates and times, quoting, encoding, handling errors, and more. For instance, if you have a file with one data column and want to get a Series object instead of a DataFrame, then you can pass squeeze=True to read_csv(). You’ll learn later on about data compression and decompression, as well as how to skip rows and columns.
JSON Files
JSON stands for JavaScript object notation. JSON files are plaintext files used for data interchange, and humans can read them easily. They follow the ISO/IEC 21778:2017 and ECMA-404 standards and use the .json extension. Python and Pandas work well with JSON files, as Python’s json library offers built-in support for them.
You can save the data from your DataFrame to a JSON file with .to_json(). Start by creating a DataFrame object again. Use the dictionary data that holds the data about countries and then apply .to_json():
>>>>>> df=pd.DataFrame(data=data).T>>> df.to_json('data-columns.json') This code produces the file data-columns.json. You can expand the code block below to see how this file should look:
{"COUNTRY":{"CHN":"China","IND":"India","USA":"US","IDN":"Indonesia","BRA":"Brazil","PAK":"Pakistan","NGA":"Nigeria","BGD":"Bangladesh","RUS":"Russia","MEX":"Mexico","JPN":"Japan","DEU":"Germany","FRA":"France","GBR":"UK","ITA":"Italy","ARG":"Argentina","DZA":"Algeria","CAN":"Canada","AUS":"Australia","KAZ":"Kazakhstan"},"POP":{"CHN":1398.72,"IND":1351.16,"USA":329.74,"IDN":268.07,"BRA":210.32,"PAK":205.71,"NGA":200.96,"BGD":167.09,"RUS":146.79,"MEX":126.58,"JPN":126.22,"DEU":83.02,"FRA":67.02,"GBR":66.44,"ITA":60.36,"ARG":44.94,"DZA":43.38,"CAN":37.59,"AUS":25.47,"KAZ":18.53},"AREA":{"CHN":9596.96,"IND":3287.26,"USA":9833.52,"IDN":1910.93,"BRA":8515.77,"PAK":881.91,"NGA":923.77,"BGD":147.57,"RUS":17098.25,"MEX":1964.38,"JPN":377.97,"DEU":357.11,"FRA":640.68,"GBR":242.5,"ITA":301.34,"ARG":2780.4,"DZA":2381.74,"CAN":9984.67,"AUS":7692.02,"KAZ":2724.9},"GDP":{"CHN":12234.78,"IND":2575.67,"USA":19485.39,"IDN":1015.54,"BRA":2055.51,"PAK":302.14,"NGA":375.77,"BGD":245.63,"RUS":1530.75,"MEX":1158.23,"JPN":4872.42,"DEU":3693.2,"FRA":2582.49,"GBR":2631.23,"ITA":1943.84,"ARG":637.49,"DZA":167.56,"CAN":1647.12,"AUS":1408.68,"KAZ":159.41},"CONT":{"CHN":"Asia","IND":"Asia","USA":"N.America","IDN":"Asia","BRA":"S.America","PAK":"Asia","NGA":"Africa","BGD":"Asia","RUS":null,"MEX":"N.America","JPN":"Asia","DEU":"Europe","FRA":"Europe","GBR":"Europe","ITA":"Europe","ARG":"S.America","DZA":"Africa","CAN":"N.America","AUS":"Oceania","KAZ":"Asia"},"IND_DAY":{"CHN":null,"IND":"1947-08-15","USA":"1776-07-04","IDN":"1945-08-17","BRA":"1822-09-07","PAK":"1947-08-14","NGA":"1960-10-01","BGD":"1971-03-26","RUS":"1992-06-12","MEX":"1810-09-16","JPN":null,"DEU":null,"FRA":"1789-07-14","GBR":null,"ITA":null,"ARG":"1816-07-09","DZA":"1962-07-05","CAN":"1867-07-01","AUS":null,"KAZ":"1991-12-16"}}data-columns.json has one large dictionary with the column labels as keys and the corresponding inner dictionaries as values.
You can get a different file structure if you pass an argument for the optional parameter orient:
>>>>>> df.to_json('data-index.json',orient='index') The orient parameter defaults to 'columns'. Here, you’ve set it to index.
You should get a new file data-index.json. You can expand the code block below to see the changes:
{"CHN":{"COUNTRY":"China","POP":1398.72,"AREA":9596.96,"GDP":12234.78,"CONT":"Asia","IND_DAY":null},"IND":{"COUNTRY":"India","POP":1351.16,"AREA":3287.26,"GDP":2575.67,"CONT":"Asia","IND_DAY":"1947-08-15"},"USA":{"COUNTRY":"US","POP":329.74,"AREA":9833.52,"GDP":19485.39,"CONT":"N.America","IND_DAY":"1776-07-04"},"IDN":{"COUNTRY":"Indonesia","POP":268.07,"AREA":1910.93,"GDP":1015.54,"CONT":"Asia","IND_DAY":"1945-08-17"},"BRA":{"COUNTRY":"Brazil","POP":210.32,"AREA":8515.77,"GDP":2055.51,"CONT":"S.America","IND_DAY":"1822-09-07"},"PAK":{"COUNTRY":"Pakistan","POP":205.71,"AREA":881.91,"GDP":302.14,"CONT":"Asia","IND_DAY":"1947-08-14"},"NGA":{"COUNTRY":"Nigeria","POP":200.96,"AREA":923.77,"GDP":375.77,"CONT":"Africa","IND_DAY":"1960-10-01"},"BGD":{"COUNTRY":"Bangladesh","POP":167.09,"AREA":147.57,"GDP":245.63,"CONT":"Asia","IND_DAY":"1971-03-26"},"RUS":{"COUNTRY":"Russia","POP":146.79,"AREA":17098.25,"GDP":1530.75,"CONT":null,"IND_DAY":"1992-06-12"},"MEX":{"COUNTRY":"Mexico","POP":126.58,"AREA":1964.38,"GDP":1158.23,"CONT":"N.America","IND_DAY":"1810-09-16"},"JPN":{"COUNTRY":"Japan","POP":126.22,"AREA":377.97,"GDP":4872.42,"CONT":"Asia","IND_DAY":null},"DEU":{"COUNTRY":"Germany","POP":83.02,"AREA":357.11,"GDP":3693.2,"CONT":"Europe","IND_DAY":null},"FRA":{"COUNTRY":"France","POP":67.02,"AREA":640.68,"GDP":2582.49,"CONT":"Europe","IND_DAY":"1789-07-14"},"GBR":{"COUNTRY":"UK","POP":66.44,"AREA":242.5,"GDP":2631.23,"CONT":"Europe","IND_DAY":null},"ITA":{"COUNTRY":"Italy","POP":60.36,"AREA":301.34,"GDP":1943.84,"CONT":"Europe","IND_DAY":null},"ARG":{"COUNTRY":"Argentina","POP":44.94,"AREA":2780.4,"GDP":637.49,"CONT":"S.America","IND_DAY":"1816-07-09"},"DZA":{"COUNTRY":"Algeria","POP":43.38,"AREA":2381.74,"GDP":167.56,"CONT":"Africa","IND_DAY":"1962-07-05"},"CAN":{"COUNTRY":"Canada","POP":37.59,"AREA":9984.67,"GDP":1647.12,"CONT":"N.America","IND_DAY":"1867-07-01"},"AUS":{"COUNTRY":"Australia","POP":25.47,"AREA":7692.02,"GDP":1408.68,"CONT":"Oceania","IND_DAY":null},"KAZ":{"COUNTRY":"Kazakhstan","POP":18.53,"AREA":2724.9,"GDP":159.41,"CONT":"Asia","IND_DAY":"1991-12-16"}}data-index.json also has one large dictionary, but this time the row labels are the keys, and the inner dictionaries are the values.
There are few more options for orient. One of them is 'records':
>>>>>> df.to_json('data-records.json',orient='records') This code should yield the file data-records.json. You can expand the code block below to see the content:
[{"COUNTRY":"China","POP":1398.72,"AREA":9596.96,"GDP":12234.78,"CONT":"Asia","IND_DAY":null},{"COUNTRY":"India","POP":1351.16,"AREA":3287.26,"GDP":2575.67,"CONT":"Asia","IND_DAY":"1947-08-15"},{"COUNTRY":"US","POP":329.74,"AREA":9833.52,"GDP":19485.39,"CONT":"N.America","IND_DAY":"1776-07-04"},{"COUNTRY":"Indonesia","POP":268.07,"AREA":1910.93,"GDP":1015.54,"CONT":"Asia","IND_DAY":"1945-08-17"},{"COUNTRY":"Brazil","POP":210.32,"AREA":8515.77,"GDP":2055.51,"CONT":"S.America","IND_DAY":"1822-09-07"},{"COUNTRY":"Pakistan","POP":205.71,"AREA":881.91,"GDP":302.14,"CONT":"Asia","IND_DAY":"1947-08-14"},{"COUNTRY":"Nigeria","POP":200.96,"AREA":923.77,"GDP":375.77,"CONT":"Africa","IND_DAY":"1960-10-01"},{"COUNTRY":"Bangladesh","POP":167.09,"AREA":147.57,"GDP":245.63,"CONT":"Asia","IND_DAY":"1971-03-26"},{"COUNTRY":"Russia","POP":146.79,"AREA":17098.25,"GDP":1530.75,"CONT":null,"IND_DAY":"1992-06-12"},{"COUNTRY":"Mexico","POP":126.58,"AREA":1964.38,"GDP":1158.23,"CONT":"N.America","IND_DAY":"1810-09-16"},{"COUNTRY":"Japan","POP":126.22,"AREA":377.97,"GDP":4872.42,"CONT":"Asia","IND_DAY":null},{"COUNTRY":"Germany","POP":83.02,"AREA":357.11,"GDP":3693.2,"CONT":"Europe","IND_DAY":null},{"COUNTRY":"France","POP":67.02,"AREA":640.68,"GDP":2582.49,"CONT":"Europe","IND_DAY":"1789-07-14"},{"COUNTRY":"UK","POP":66.44,"AREA":242.5,"GDP":2631.23,"CONT":"Europe","IND_DAY":null},{"COUNTRY":"Italy","POP":60.36,"AREA":301.34,"GDP":1943.84,"CONT":"Europe","IND_DAY":null},{"COUNTRY":"Argentina","POP":44.94,"AREA":2780.4,"GDP":637.49,"CONT":"S.America","IND_DAY":"1816-07-09"},{"COUNTRY":"Algeria","POP":43.38,"AREA":2381.74,"GDP":167.56,"CONT":"Africa","IND_DAY":"1962-07-05"},{"COUNTRY":"Canada","POP":37.59,"AREA":9984.67,"GDP":1647.12,"CONT":"N.America","IND_DAY":"1867-07-01"},{"COUNTRY":"Australia","POP":25.47,"AREA":7692.02,"GDP":1408.68,"CONT":"Oceania","IND_DAY":null},{"COUNTRY":"Kazakhstan","POP":18.53,"AREA":2724.9,"GDP":159.41,"CONT":"Asia","IND_DAY":"1991-12-16"}]data-records.json holds a list with one dictionary for each row. The row labels are not written.
You can get another interesting file structure with orient='split':
>>>>>> df.to_json('data-split.json',orient='split') The resulting file is data-split.json. You can expand the code block below to see how this file should look:
{"columns":["COUNTRY","POP","AREA","GDP","CONT","IND_DAY"],"index":["CHN","IND","USA","IDN","BRA","PAK","NGA","BGD","RUS","MEX","JPN","DEU","FRA","GBR","ITA","ARG","DZA","CAN","AUS","KAZ"],"data":[["China",1398.72,9596.96,12234.78,"Asia",null],["India",1351.16,3287.26,2575.67,"Asia","1947-08-15"],["US",329.74,9833.52,19485.39,"N.America","1776-07-04"],["Indonesia",268.07,1910.93,1015.54,"Asia","1945-08-17"],["Brazil",210.32,8515.77,2055.51,"S.America","1822-09-07"],["Pakistan",205.71,881.91,302.14,"Asia","1947-08-14"],["Nigeria",200.96,923.77,375.77,"Africa","1960-10-01"],["Bangladesh",167.09,147.57,245.63,"Asia","1971-03-26"],["Russia",146.79,17098.25,1530.75,null,"1992-06-12"],["Mexico",126.58,1964.38,1158.23,"N.America","1810-09-16"],["Japan",126.22,377.97,4872.42,"Asia",null],["Germany",83.02,357.11,3693.2,"Europe",null],["France",67.02,640.68,2582.49,"Europe","1789-07-14"],["UK",66.44,242.5,2631.23,"Europe",null],["Italy",60.36,301.34,1943.84,"Europe",null],["Argentina",44.94,2780.4,637.49,"S.America","1816-07-09"],["Algeria",43.38,2381.74,167.56,"Africa","1962-07-05"],["Canada",37.59,9984.67,1647.12,"N.America","1867-07-01"],["Australia",25.47,7692.02,1408.68,"Oceania",null],["Kazakhstan",18.53,2724.9,159.41,"Asia","1991-12-16"]]}data-split.json contains one dictionary that holds the following lists:
- The names of the columns
- The labels of the rows
- The inner lists (two-dimensional sequence) that hold data values
If you don’t provide the value for the optional parameter path_or_buf that defines the file path, then .to_json() will return a JSON string instead of writing the results to a file. This behavior is consistent with .to_csv().
There are other optional parameters you can use. For instance, you can set index=False to forego saving row labels. You can manipulate precision with double_precision, and dates with date_format and date_unit. These last two parameters are particularly important when you have time series among your data:
>>>>>> df=pd.DataFrame(data=data).T>>> df['IND_DAY']=pd.to_datetime(df['IND_DAY'])>>> df.dtypesCOUNTRY objectPOP objectAREA objectGDP objectCONT objectIND_DAY datetime64[ns]dtype: object>>> df.to_json('data-time.json') In this example, you’ve created the DataFrame from the dictionary data and used to_datetime() to convert the values in the last column to datetime64. You can expand the code block below to see the resulting file:
{"COUNTRY":{"CHN":"China","IND":"India","USA":"US","IDN":"Indonesia","BRA":"Brazil","PAK":"Pakistan","NGA":"Nigeria","BGD":"Bangladesh","RUS":"Russia","MEX":"Mexico","JPN":"Japan","DEU":"Germany","FRA":"France","GBR":"UK","ITA":"Italy","ARG":"Argentina","DZA":"Algeria","CAN":"Canada","AUS":"Australia","KAZ":"Kazakhstan"},"POP":{"CHN":1398.72,"IND":1351.16,"USA":329.74,"IDN":268.07,"BRA":210.32,"PAK":205.71,"NGA":200.96,"BGD":167.09,"RUS":146.79,"MEX":126.58,"JPN":126.22,"DEU":83.02,"FRA":67.02,"GBR":66.44,"ITA":60.36,"ARG":44.94,"DZA":43.38,"CAN":37.59,"AUS":25.47,"KAZ":18.53},"AREA":{"CHN":9596.96,"IND":3287.26,"USA":9833.52,"IDN":1910.93,"BRA":8515.77,"PAK":881.91,"NGA":923.77,"BGD":147.57,"RUS":17098.25,"MEX":1964.38,"JPN":377.97,"DEU":357.11,"FRA":640.68,"GBR":242.5,"ITA":301.34,"ARG":2780.4,"DZA":2381.74,"CAN":9984.67,"AUS":7692.02,"KAZ":2724.9},"GDP":{"CHN":12234.78,"IND":2575.67,"USA":19485.39,"IDN":1015.54,"BRA":2055.51,"PAK":302.14,"NGA":375.77,"BGD":245.63,"RUS":1530.75,"MEX":1158.23,"JPN":4872.42,"DEU":3693.2,"FRA":2582.49,"GBR":2631.23,"ITA":1943.84,"ARG":637.49,"DZA":167.56,"CAN":1647.12,"AUS":1408.68,"KAZ":159.41},"CONT":{"CHN":"Asia","IND":"Asia","USA":"N.America","IDN":"Asia","BRA":"S.America","PAK":"Asia","NGA":"Africa","BGD":"Asia","RUS":null,"MEX":"N.America","JPN":"Asia","DEU":"Europe","FRA":"Europe","GBR":"Europe","ITA":"Europe","ARG":"S.America","DZA":"Africa","CAN":"N.America","AUS":"Oceania","KAZ":"Asia"},"IND_DAY":{"CHN":null,"IND":-706320000000,"USA":-6106060800000,"IDN":-769219200000,"BRA":-4648924800000,"PAK":-706406400000,"NGA":-291945600000,"BGD":38793600000,"RUS":708307200000,"MEX":-5026838400000,"JPN":null,"DEU":null,"FRA":-5694969600000,"GBR":null,"ITA":null,"ARG":-4843411200000,"DZA":-236476800000,"CAN":-3234729600000,"AUS":null,"KAZ":692841600000}}In this file, you have large integers instead of dates for the independence days. That’s because the default value of the optional parameter date_format is 'epoch' whenever orient isn’t 'table'. This default behavior expresses dates as an epoch in milliseconds relative to midnight on January 1, 1970.
However, if you pass date_format='iso', then you’ll get the dates in the ISO 8601 format. In addition, date_unit decides the units of time:
>>>>>> df=pd.DataFrame(data=data).T>>> df['IND_DAY']=pd.to_datetime(df['IND_DAY'])>>> df.to_json('new-data-time.json',date_format='iso',date_unit='s') This code produces the following JSON file:
{"COUNTRY":{"CHN":"China","IND":"India","USA":"US","IDN":"Indonesia","BRA":"Brazil","PAK":"Pakistan","NGA":"Nigeria","BGD":"Bangladesh","RUS":"Russia","MEX":"Mexico","JPN":"Japan","DEU":"Germany","FRA":"France","GBR":"UK","ITA":"Italy","ARG":"Argentina","DZA":"Algeria","CAN":"Canada","AUS":"Australia","KAZ":"Kazakhstan"},"POP":{"CHN":1398.72,"IND":1351.16,"USA":329.74,"IDN":268.07,"BRA":210.32,"PAK":205.71,"NGA":200.96,"BGD":167.09,"RUS":146.79,"MEX":126.58,"JPN":126.22,"DEU":83.02,"FRA":67.02,"GBR":66.44,"ITA":60.36,"ARG":44.94,"DZA":43.38,"CAN":37.59,"AUS":25.47,"KAZ":18.53},"AREA":{"CHN":9596.96,"IND":3287.26,"USA":9833.52,"IDN":1910.93,"BRA":8515.77,"PAK":881.91,"NGA":923.77,"BGD":147.57,"RUS":17098.25,"MEX":1964.38,"JPN":377.97,"DEU":357.11,"FRA":640.68,"GBR":242.5,"ITA":301.34,"ARG":2780.4,"DZA":2381.74,"CAN":9984.67,"AUS":7692.02,"KAZ":2724.9},"GDP":{"CHN":12234.78,"IND":2575.67,"USA":19485.39,"IDN":1015.54,"BRA":2055.51,"PAK":302.14,"NGA":375.77,"BGD":245.63,"RUS":1530.75,"MEX":1158.23,"JPN":4872.42,"DEU":3693.2,"FRA":2582.49,"GBR":2631.23,"ITA":1943.84,"ARG":637.49,"DZA":167.56,"CAN":1647.12,"AUS":1408.68,"KAZ":159.41},"CONT":{"CHN":"Asia","IND":"Asia","USA":"N.America","IDN":"Asia","BRA":"S.America","PAK":"Asia","NGA":"Africa","BGD":"Asia","RUS":null,"MEX":"N.America","JPN":"Asia","DEU":"Europe","FRA":"Europe","GBR":"Europe","ITA":"Europe","ARG":"S.America","DZA":"Africa","CAN":"N.America","AUS":"Oceania","KAZ":"Asia"},"IND_DAY":{"CHN":null,"IND":"1947-08-15T00:00:00Z","USA":"1776-07-04T00:00:00Z","IDN":"1945-08-17T00:00:00Z","BRA":"1822-09-07T00:00:00Z","PAK":"1947-08-14T00:00:00Z","NGA":"1960-10-01T00:00:00Z","BGD":"1971-03-26T00:00:00Z","RUS":"1992-06-12T00:00:00Z","MEX":"1810-09-16T00:00:00Z","JPN":null,"DEU":null,"FRA":"1789-07-14T00:00:00Z","GBR":null,"ITA":null,"ARG":"1816-07-09T00:00:00Z","DZA":"1962-07-05T00:00:00Z","CAN":"1867-07-01T00:00:00Z","AUS":null,"KAZ":"1991-12-16T00:00:00Z"}}The dates in the resulting file are in the ISO 8601 format.
You can load the data from a JSON file with read_json():
>>>>>> df=pd.read_json('data-index.json',orient='index',... convert_dates=['IND_DAY']) The parameter convert_dates has a similar purpose as parse_dates when you use it to read CSV files. The optional parameter orient is very important because it specifies how Pandas understands the structure of the file.
There are other optional parameters you can use as well:
- Set the encoding with
encoding. - Manipulate dates with
convert_dates and keep_default_dates. - Impact precision with
dtype and precise_float. - Decode numeric data directly to NumPy arrays with
numpy=True.
Note that you might lose the order of rows and columns when using the JSON format to store your data.
HTML Files
An HTML is a plaintext file that uses hypertext markup language to help browsers render web pages. The extensions for HTML files are .html and .htm. You’ll need to install an HTML parser library like lxml or html5lib to be able to work with HTML files:
$pip install lxml html5lib
You can also use Conda to install the same packages:
$ conda install lxml html5lib
Once you have these libraries, you can save the contents of your DataFrame as an HTML file with .to_html():
>>>df = pd.DataFrame(data=data).Tdf.to_html('data.html') This code generates a file data.html. You can expand the code block below to see how this file should look:
<tableborder="1"class="dataframe"><thead><trstyle="text-align: right;"><th></th><th>COUNTRY</th><th>POP</th><th>AREA</th><th>GDP</th><th>CONT</th><th>IND_DAY</th></tr></thead><tbody><tr><th>CHN</th><td>China</td><td>1398.72</td><td>9596.96</td><td>12234.8</td><td>Asia</td><td>NaN</td></tr><tr><th>IND</th><td>India</td><td>1351.16</td><td>3287.26</td><td>2575.67</td><td>Asia</td><td>1947-08-15</td></tr><tr><th>USA</th><td>US</td><td>329.74</td><td>9833.52</td><td>19485.4</td><td>N.America</td><td>1776-07-04</td></tr><tr><th>IDN</th><td>Indonesia</td><td>268.07</td><td>1910.93</td><td>1015.54</td><td>Asia</td><td>1945-08-17</td></tr><tr><th>BRA</th><td>Brazil</td><td>210.32</td><td>8515.77</td><td>2055.51</td><td>S.America</td><td>1822-09-07</td></tr><tr><th>PAK</th><td>Pakistan</td><td>205.71</td><td>881.91</td><td>302.14</td><td>Asia</td><td>1947-08-14</td></tr><tr><th>NGA</th><td>Nigeria</td><td>200.96</td><td>923.77</td><td>375.77</td><td>Africa</td><td>1960-10-01</td></tr><tr><th>BGD</th><td>Bangladesh</td><td>167.09</td><td>147.57</td><td>245.63</td><td>Asia</td><td>1971-03-26</td></tr><tr><th>RUS</th><td>Russia</td><td>146.79</td><td>17098.2</td><td>1530.75</td><td>NaN</td><td>1992-06-12</td></tr><tr><th>MEX</th><td>Mexico</td><td>126.58</td><td>1964.38</td><td>1158.23</td><td>N.America</td><td>1810-09-16</td></tr><tr><th>JPN</th><td>Japan</td><td>126.22</td><td>377.97</td><td>4872.42</td><td>Asia</td><td>NaN</td></tr><tr><th>DEU</th><td>Germany</td><td>83.02</td><td>357.11</td><td>3693.2</td><td>Europe</td><td>NaN</td></tr><tr><th>FRA</th><td>France</td><td>67.02</td><td>640.68</td><td>2582.49</td><td>Europe</td><td>1789-07-14</td></tr><tr><th>GBR</th><td>UK</td><td>66.44</td><td>242.5</td><td>2631.23</td><td>Europe</td><td>NaN</td></tr><tr><th>ITA</th><td>Italy</td><td>60.36</td><td>301.34</td><td>1943.84</td><td>Europe</td><td>NaN</td></tr><tr><th>ARG</th><td>Argentina</td><td>44.94</td><td>2780.4</td><td>637.49</td><td>S.America</td><td>1816-07-09</td></tr><tr><th>DZA</th><td>Algeria</td><td>43.38</td><td>2381.74</td><td>167.56</td><td>Africa</td><td>1962-07-05</td></tr><tr><th>CAN</th><td>Canada</td><td>37.59</td><td>9984.67</td><td>1647.12</td><td>N.America</td><td>1867-07-01</td></tr><tr><th>AUS</th><td>Australia</td><td>25.47</td><td>7692.02</td><td>1408.68</td><td>Oceania</td><td>NaN</td></tr><tr><th>KAZ</th><td>Kazakhstan</td><td>18.53</td><td>2724.9</td><td>159.41</td><td>Asia</td><td>1991-12-16</td></tr></tbody></table>
This file shows the DataFrame contents nicely. However, notice that you haven’t obtained an entire web page. You’ve just output the data that corresponds to df in the HTML format.
.to_html() won’t create a file if you don’t provide the optional parameter buf, which denotes the buffer to write to. If you leave this parameter out, then your code will return a string as it did with .to_csv() and .to_json().
Here are some other optional parameters:
header determines whether to save the column names.index determines whether to save the row labels.classes assigns cascading style sheet (CSS) classes.render_links specifies whether to convert URLs to HTML links.table_id assigns the CSS id to the table tag.escape decides whether to convert the characters <, >, and & to HTML-safe strings.
You use parameters like these to specify different aspects of the resulting files or strings.
You can create a DataFrame object from a suitable HTML file using read_html(), which will return a DataFrame instance or a list of them:
>>>>>> df=pd.read_html('data.html',index_col=0,parse_dates=['IND_DAY']) This is very similar to what you did when reading CSV files. You also have parameters that help you work with dates, missing values, precision, encoding, HTML parsers, and more.
Excel Files
You’ve already learned how to read and write Excel files with Pandas. However, there are a few more options worth considering. For one, when you use .to_excel(), you can specify the name of the target worksheet with the optional parameter sheet_name:
>>>>>> df=pd.DataFrame(data=data).T>>> df.to_excel('data.xlsx',sheet_name='COUNTRIES') Here, you create a file data.xlsx with a worksheet called COUNTRIES that stores the data. The string 'data.xlsx' is the argument for the parameter excel_writer that defines the name of the Excel file or its path.
The optional parameters startrow and startcol both default to 0 and indicate the upper left-most cell where the data should start being written:
>>>>>> df.to_excel('data-shifted.xlsx',sheet_name='COUNTRIES',... startrow=2,startcol=4) Here, you specify that the table should start in the third row and the fifth column. You also used zero-based indexing, so the third row is denoted by 2 and the fifth column by 4.
Now the resulting worksheet looks like this:
![mmst-pandas-rw-files-excel-shifted]()
As you can see, the table starts in the third row 2 and the fifth column E.
.read_excel() also has the optional parameter sheet_name that specifies which worksheets to read when loading data. It can take on one of the following values:
- The zero-based index of the worksheet
- The name of the worksheet
- The list of indices or names to read multiple sheets
- The value
None to read all sheets
Here’s how you would use this parameter in your code:
>>>>>> df=pd.read_excel('data.xlsx',sheet_name=0,index_col=0,... parse_dates=['IND_DAY'])>>> df=pd.read_excel('data.xlsx',sheet_name='COUNTRIES',index_col=0,... parse_dates=['IND_DAY']) Both statements above create the same DataFrame because the sheet_name parameters have the same values. In both cases, sheet_name=0 and sheet_name='COUNTRIES' refer to the same worksheet. The argument parse_dates=['IND_DAY'] tells Pandas to try to consider the values in this column as dates or times.
There are other optional parameters you can use with .read_excel() and .to_excel() to determine the Excel engine, the encoding, the way to handle missing values and infinities, the method for writing column names and row labels, and so on.
SQL Files
Pandas IO tools can also read and write databases. In this next example, you’ll write your data to a database called data.db. To get started, you’ll need the SQLAlchemy package. To learn more about it, you can read the official ORM tutorial. You’ll also need the database driver. Python has a built-in driver for SQLite.
You can install SQLAlchemy with pip:
You can also install it with Conda:
$ conda install sqlalchemy
Once you have SQLAlchemy installed, import create_engine() and create a database engine:
>>>>>> fromsqlalchemyimportcreate_engine>>> engine=create_engine('sqlite:///data.db',echo=False) Now that you have everything set up, the next step is to create a DataFrame object. It’s convenient to specify the data types and apply .to_sql().
>>>>>> dtypes={'POP':'float64','AREA':'float64','GDP':'float64',... 'IND_DAY':'datetime64'}>>> df=pd.DataFrame(data=data).T.astype(dtype=dtypes)>>> df.dtypesCOUNTRY objectPOP float64AREA float64GDP float64CONT objectIND_DAY datetime64[ns]dtype: object .astype() is a very convenient method you can use to set multiple data types at once.
Once you’ve created your DataFrame, you can save it to the database with .to_sql():
>>>>>> df.to_sql('data.db',con=engine,index_label='ID') The parameter con is used to specify the database connection or engine that you want to use. The optional parameter index_label specifies how to call the database column with the row labels. You’ll often see it take on the value ID, Id, or id.
You should get the database data.db with a single table that looks like this:
![mmst-pandas-rw-files-db]()
The first column contains the row labels. To omit writing them into the database, pass index=False to .to_sql(). The other columns correspond to the columns of the DataFrame.
There are a few more optional parameters. For example, you can use schema to specify the database schema and dtype to determine the types of the database columns. You can also use if_exists, which says what to do if a database with the same name and path already exists:
if_exists='fail' raises a ValueError and is the default.if_exists='replace' drops the table and inserts new values.if_exists='append' inserts new values into the table.
You can load the data from the database with read_sql():
>>>>>> df=pd.read_sql('data.db',con=engine,index_col='ID')>>> df COUNTRY POP AREA GDP CONT IND_DAYIDCHN China 1398.72 9596.96 12234.78 Asia NaTIND India 1351.16 3287.26 2575.67 Asia 1947-08-15USA US 329.74 9833.52 19485.39 N.America 1776-07-04IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26RUS Russia 146.79 17098.25 1530.75 None 1992-06-12MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16JPN Japan 126.22 377.97 4872.42 Asia NaTDEU Germany 83.02 357.11 3693.20 Europe NaTFRA France 67.02 640.68 2582.49 Europe 1789-07-14GBR UK 66.44 242.50 2631.23 Europe NaTITA Italy 60.36 301.34 1943.84 Europe NaTARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09DZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01AUS Australia 25.47 7692.02 1408.68 Oceania NaTKAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16 The parameter index_col specifies the name of the column with the row labels. Note that this inserts an extra row after the header that starts with ID. You can fix this behavior with the following line of code:
>>>>>> df.index.name=None>>> df COUNTRY POP AREA GDP CONT IND_DAYCHN China 1398.72 9596.96 12234.78 Asia NaTIND India 1351.16 3287.26 2575.67 Asia 1947-08-15USA US 329.74 9833.52 19485.39 N.America 1776-07-04IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26RUS Russia 146.79 17098.25 1530.75 None 1992-06-12MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16JPN Japan 126.22 377.97 4872.42 Asia NaTDEU Germany 83.02 357.11 3693.20 Europe NaTFRA France 67.02 640.68 2582.49 Europe 1789-07-14GBR UK 66.44 242.50 2631.23 Europe NaTITA Italy 60.36 301.34 1943.84 Europe NaTARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09DZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01AUS Australia 25.47 7692.02 1408.68 Oceania NaTKAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16
Now you have the same DataFrame object as before.
Note that the continent for Russia is now None instead of nan. If you want to fill the missing values with nan, then you can use .fillna():
>>>>>> df.fillna(value=float('nan'),inplace=True) .fillna() replaces all missing values with whatever you pass to value. Here, you passed float('nan'), which says to fill all missing values with nan.
Also note that you didn’t have to pass parse_dates=['IND_DAY'] to read_sql(). That’s because your database was able to detect that the last column contains dates. However, you can pass parse_dates if you’d like. You’ll get the same results.
There are other functions that you can use to read databases, like read_sql_table() and read_sql_query(). Feel free to try them out!
Pickle Files
Pickling is the act of converting Python objects into byte streams. Unpickling is the inverse process. Python pickle files are the binary files that keep the data and hierarchy of Python objects. They usually have the extension .pickle or .pkl.
You can save your DataFrame in a pickle file with .to_pickle():
>>>>>> dtypes={'POP':'float64','AREA':'float64','GDP':'float64',... 'IND_DAY':'datetime64'}>>> df=pd.DataFrame(data=data).T.astype(dtype=dtypes)>>> df.to_pickle('data.pickle') Like you did with databases, it can be convenient first to specify the data types. Then, you create a file data.pickle to contain your data. You could also pass an integer value to the optional parameter protocol, which specifies the protocol of the pickler.
You can get the data from a pickle file with read_pickle():
>>>>>> df=pd.read_pickle('data.pickle')>>> df COUNTRY POP AREA GDP CONT IND_DAYCHN China 1398.72 9596.96 12234.78 Asia NaTIND India 1351.16 3287.26 2575.67 Asia 1947-08-15USA US 329.74 9833.52 19485.39 N.America 1776-07-04IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26RUS Russia 146.79 17098.25 1530.75 NaN 1992-06-12MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16JPN Japan 126.22 377.97 4872.42 Asia NaTDEU Germany 83.02 357.11 3693.20 Europe NaTFRA France 67.02 640.68 2582.49 Europe 1789-07-14GBR UK 66.44 242.50 2631.23 Europe NaTITA Italy 60.36 301.34 1943.84 Europe NaTARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09DZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01AUS Australia 25.47 7692.02 1408.68 Oceania NaTKAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16 read_pickle() returns the DataFrame with the stored data. You can also check the data types:
>>>>>> df.dtypesCOUNTRY objectPOP float64AREA float64GDP float64CONT objectIND_DAY datetime64[ns]dtype: object
These are the same ones that you specified before using .to_pickle().
As a word of caution, you should always beware of loading pickles from untrusted sources. This can be dangerous! When you unpickle an untrustworthy file, it could execute arbitrary code on your machine, gain remote access to your computer, or otherwise exploit your device in other ways.
Working With Big Data
If your files are too large for saving or processing, then there are several approaches you can take to reduce the required disk space:
- Compress your files
- Choose only the columns you want
- Omit the rows you don’t need
- Force the use of less precise data types
- Split the data into chunks
You’ll take a look at each of these techniques in turn.
Compress and Decompress Files
You can create an archive file like you would a regular one, with the addition of a suffix that corresponds to the desired compression type:
Pandas can deduce the compression type by itself:
>>>>>> df=pd.DataFrame(data=data).T>>> df.to_csv('data.csv.zip') Here, you create a compressed .csv file as an archive. The size of the regular .csv file is 1048 bytes, while the compressed file only has 766 bytes.
You can open this compressed file as usual with the Pandas read_csv() function:
>>>>>> df=pd.read_csv('data.csv.zip',index_col=0,... parse_dates=['IND_DAY'])>>> df COUNTRY POP AREA GDP CONT IND_DAYCHN China 1398.72 9596.96 12234.78 Asia NaTIND India 1351.16 3287.26 2575.67 Asia 1947-08-15USA US 329.74 9833.52 19485.39 N.America 1776-07-04IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26RUS Russia 146.79 17098.25 1530.75 NaN 1992-06-12MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16JPN Japan 126.22 377.97 4872.42 Asia NaTDEU Germany 83.02 357.11 3693.20 Europe NaTFRA France 67.02 640.68 2582.49 Europe 1789-07-14GBR UK 66.44 242.50 2631.23 Europe NaTITA Italy 60.36 301.34 1943.84 Europe NaTARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09DZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01AUS Australia 25.47 7692.02 1408.68 Oceania NaTKAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16 read_csv() decompresses the file before reading it into a DataFrame.
You can specify the type of compression with the optional parameter compression, which can take on any of the following values:
'infer''gzip''bz2''zip''xz'None
The default value compression='infer' indicates that Pandas should deduce the compression type from the file extension.
Here’s how you would compress a pickle file:
>>>>>> df=pd.DataFrame(data=data).T>>> df.to_pickle('data.pickle.compress',compression='gzip') You should get the file data.pickle.compress that you can later decompress and read:
>>>>>> df=pd.read_pickle('data.pickle.compress',compression='gzip') df again corresponds to the DataFrame with the same data as before.
You can give the other compression methods a try, as well. If you’re using pickle files, then keep in mind that the .zip format supports reading only.
Choose Columns
The Pandas read_csv() and read_excel() functions have the optional parameter usecols that you can use to specify the columns you want to load from the file. You can pass the list of column names as the corresponding argument:
>>>>>> df=pd.read_csv('data.csv',usecols=['COUNTRY','AREA'])>>> df COUNTRY AREA0 China 9596.961 India 3287.262 US 9833.523 Indonesia 1910.934 Brazil 8515.775 Pakistan 881.916 Nigeria 923.777 Bangladesh 147.578 Russia 17098.259 Mexico 1964.3810 Japan 377.9711 Germany 357.1112 France 640.6813 UK 242.5014 Italy 301.3415 Argentina 2780.4016 Algeria 2381.7417 Canada 9984.6718 Australia 7692.0219 Kazakhstan 2724.90 Now you have a DataFrame that contains less data than before. Here, there are only the names of the countries and their areas.
Instead of the column names, you can also pass their indices:
>>>>>> df=pd.read_csv('data.csv',index_col=0,usecols=[0,1,3])>>> df COUNTRY AREACHN China 9596.96IND India 3287.26USA US 9833.52IDN Indonesia 1910.93BRA Brazil 8515.77PAK Pakistan 881.91NGA Nigeria 923.77BGD Bangladesh 147.57RUS Russia 17098.25MEX Mexico 1964.38JPN Japan 377.97DEU Germany 357.11FRA France 640.68GBR UK 242.50ITA Italy 301.34ARG Argentina 2780.40DZA Algeria 2381.74CAN Canada 9984.67AUS Australia 7692.02KAZ Kazakhstan 2724.90 Expand the code block below to compare these results with the file 'data.csv':
,COUNTRY,POP,AREA,GDP,CONT,IND_DAYCHN,China,1398.72,9596.96,12234.78,Asia,IND,India,1351.16,3287.26,2575.67,Asia,1947-08-15USA,US,329.74,9833.52,19485.39,N.America,1776-07-04IDN,Indonesia,268.07,1910.93,1015.54,Asia,1945-08-17BRA,Brazil,210.32,8515.77,2055.51,S.America,1822-09-07PAK,Pakistan,205.71,881.91,302.14,Asia,1947-08-14NGA,Nigeria,200.96,923.77,375.77,Africa,1960-10-01BGD,Bangladesh,167.09,147.57,245.63,Asia,1971-03-26RUS,Russia,146.79,17098.25,1530.75,,1992-06-12MEX,Mexico,126.58,1964.38,1158.23,N.America,1810-09-16JPN,Japan,126.22,377.97,4872.42,Asia,DEU,Germany,83.02,357.11,3693.2,Europe,FRA,France,67.02,640.68,2582.49,Europe,1789-07-14GBR,UK,66.44,242.5,2631.23,Europe,ITA,Italy,60.36,301.34,1943.84,Europe,ARG,Argentina,44.94,2780.4,637.49,S.America,1816-07-09DZA,Algeria,43.38,2381.74,167.56,Africa,1962-07-05CAN,Canada,37.59,9984.67,1647.12,N.America,1867-07-01AUS,Australia,25.47,7692.02,1408.68,Oceania,KAZ,Kazakhstan,18.53,2724.9,159.41,Asia,1991-12-16
You can see the following columns:
- The column at index
0 contains the row labels. - The column at index
1 contains the country names. - The column at index
3 contains the areas.
Simlarly, read_sql() has the optional parameter columns that takes a list of column names to read:
>>>>>> df=pd.read_sql('data.db',con=engine,index_col='ID',... columns=['COUNTRY','AREA'])>>> df.index.name=None>>> df COUNTRY AREACHN China 9596.96IND India 3287.26USA US 9833.52IDN Indonesia 1910.93BRA Brazil 8515.77PAK Pakistan 881.91NGA Nigeria 923.77BGD Bangladesh 147.57RUS Russia 17098.25MEX Mexico 1964.38JPN Japan 377.97DEU Germany 357.11FRA France 640.68GBR UK 242.50ITA Italy 301.34ARG Argentina 2780.40DZA Algeria 2381.74CAN Canada 9984.67AUS Australia 7692.02KAZ Kazakhstan 2724.90 Again, the DataFrame only contains the columns with the names of the countries and areas. If columns is None or omitted, then all of the columns will be read, as you saw before. The default behavior is columns=None.
Omit Rows
When you test an algorithm for data processing or machine learning, you often don’t need the entire dataset. It’s convenient to load only a subset of the data to speed up the process. The Pandas read_csv() and read_excel() functions have some optional parameters that allow you to select which rows you want to load:
skiprows: either the number of rows to skip at the beginning of the file if it’s an integer, or the zero-based indices of the rows to skip if it’s a list-like objectskipfooter: the number of rows to skip at the end of the filenrows: the number of rows to read
Here’s how you would skip rows with odd zero-based indices, keeping the even ones:
>>>>>> df=pd.read_csv('data.csv',index_col=0,skiprows=range(1,20,2))>>> df COUNTRY POP AREA GDP CONT IND_DAYIND India 1351.16 3287.26 2575.67 Asia 1947-08-15IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16DEU Germany 83.02 357.11 3693.20 Europe NaNGBR UK 66.44 242.50 2631.23 Europe NaNARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01KAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16 In this example, skiprows is range(1, 20, 2) and corresponds to the values 1, 3, …, 19. The instances of the Python built-in class range behave like sequences. The first row of the file data.csv is the header row. It has the index 0, so Pandas loads it in. The second row with index 1 corresponds to the label CHN, and Pandas skips it. The third row with the index 2 and label IND is loaded, and so on.
If you want to choose rows randomly, then skiprows can be a list or NumPy array with pseudo-random numbers, obtained either with pure Python or with NumPy.
Force Less Precise Data Types
If you’re okay with less precise data types, then you can potentially save a significant amount of memory! First, get the data types with .dtypes again:
>>>>>> df=pd.read_csv('data.csv',index_col=0,parse_dates=['IND_DAY'])>>> df.dtypesCOUNTRY objectPOP float64AREA float64GDP float64CONT objectIND_DAY datetime64[ns]dtype: object The columns with the floating-point numbers are 64-bit floats. Each number of this type float64 consumes 64 bits or 8 bytes. Each column has 20 numbers and requires 160 bytes. You can verify this with .memory_usage():
>>>>>> df.memory_usage()Index 160COUNTRY 160POP 160AREA 160GDP 160CONT 160IND_DAY 160dtype: int64
.memory_usage() returns an instance of Series with the memory usage of each column in bytes. You can conveniently combine it with .loc[] and .sum() to get the memory for a group of columns:
>>>>>> df.loc[:,['POP','AREA','GDP']].memory_usage(index=False).sum()480
This example shows how you can combine the numeric columns 'POP', 'AREA', and 'GDP' to get their total memory requirement. The argument index=False excludes data for row labels from the resulting Series object. For these three columns, you’ll need 480 bytes.
You can also extract the data values in the form of a NumPy array with .to_numpy() or .values. Then, use the .nbytes attribute to get the total bytes consumed by the items of the array:
>>>>>> df.loc[:,['POP','AREA','GDP']].to_numpy().nbytes480
The result is the same 480 bytes. So, how do you save memory?
In this case, you can specify that your numeric columns 'POP', 'AREA', and 'GDP' should have the type float32. Use the optional parameter dtype to do this:
>>>>>> dtypes={'POP':'float32','AREA':'float32','GDP':'float32'}>>> df=pd.read_csv('data.csv',index_col=0,dtype=dtypes,... parse_dates=['IND_DAY']) The dictionary dtypes specifies the desired data types for each column. It’s passed to the Pandas read_csv() function as the argument that corresponds to the parameter dtype.
Now you can verify that each numeric column needs 80 bytes, or 4 bytes per item:
>>>>>> df.dtypesCOUNTRY objectPOP float32AREA float32GDP float32CONT objectIND_DAY datetime64[ns]dtype: object>>> df.memory_usage()Index 160COUNTRY 160POP 80AREA 80GDP 80CONT 160IND_DAY 160dtype: int64>>> df.loc[:,['POP','AREA','GDP']].memory_usage(index=False).sum()240>>> df.loc[:,['POP','AREA','GDP']].to_numpy().nbytes240
Each value is a floating-point number of 32 bits or 4 bytes. The three numeric columns contain 20 items each. In total, you’ll need 240 bytes of memory when you work with the type float32. This is half the size of the 480 bytes you’d need to work with float64.
In addition to saving memory, you can significantly reduce the time required to process data by using float32 instead of float64 in some cases.
Use Chunks to Iterate Through Files
Another way to deal with very large datasets is to split the data into smaller chunks and process one chunk at a time. If you use read_csv(), read_json() or read_sql(), then you can specify the optional parameter chunksize:
>>>>>> data_chunk=pd.read_csv('data.csv',index_col=0,chunksize=8)>>> type(data_chunk)<class 'pandas.io.parsers.TextFileReader'>>>> hasattr(data_chunk,'__iter__')True>>> hasattr(data_chunk,'__next__')True chunksize defaults to None and can take on an integer value that indicates the number of items in a single chunk. When chunksize is an integer, read_csv() returns an iterable that you can use in a for loop to get and process only a fragment of the dataset in each iteration:
>>>>>> fordf_chunkinpd.read_csv('data.csv',index_col=0,chunksize=8):... print(df_chunk,end='\n\n')... print('memory:',df_chunk.memory_usage().sum(),'bytes',... end='\n\n\n')... COUNTRY POP AREA GDP CONT IND_DAYCHN China 1398.72 9596.96 12234.78 Asia NaNIND India 1351.16 3287.26 2575.67 Asia 1947-08-15USA US 329.74 9833.52 19485.39 N.America 1776-07-04IDN Indonesia 268.07 1910.93 1015.54 Asia 1945-08-17BRA Brazil 210.32 8515.77 2055.51 S.America 1822-09-07PAK Pakistan 205.71 881.91 302.14 Asia 1947-08-14NGA Nigeria 200.96 923.77 375.77 Africa 1960-10-01BGD Bangladesh 167.09 147.57 245.63 Asia 1971-03-26memory: 448 bytes COUNTRY POP AREA GDP CONT IND_DAYRUS Russia 146.79 17098.25 1530.75 NaN 1992-06-12MEX Mexico 126.58 1964.38 1158.23 N.America 1810-09-16JPN Japan 126.22 377.97 4872.42 Asia NaNDEU Germany 83.02 357.11 3693.20 Europe NaNFRA France 67.02 640.68 2582.49 Europe 1789-07-14GBR UK 66.44 242.50 2631.23 Europe NaNITA Italy 60.36 301.34 1943.84 Europe NaNARG Argentina 44.94 2780.40 637.49 S.America 1816-07-09memory: 448 bytes COUNTRY POP AREA GDP CONT IND_DAYDZA Algeria 43.38 2381.74 167.56 Africa 1962-07-05CAN Canada 37.59 9984.67 1647.12 N.America 1867-07-01AUS Australia 25.47 7692.02 1408.68 Oceania NaNKAZ Kazakhstan 18.53 2724.90 159.41 Asia 1991-12-16memory: 224 bytes In this example, the chunksize is 8. The first iteration of the for loop returns a DataFrame with the first eight rows of the dataset only. The second iteration returns another DataFrame with the next eight rows. The third and last iteration returns the remaining four rows.
Note: You can also pass iterator=True to force the Pandas read_csv() function to return an iterator object instead of a DataFrame object.
In each iteration, you get and process the DataFrame with the number of rows equal to chunksize. It’s possible to have fewer rows than the value of chunksize in the last iteration. You can use this functionality to control the amount of memory required to process data and keep that amount reasonably small.
Conclusion
You now know how to save the data and labels from Pandas DataFrame objects to different kinds of files. You also know how to load your data from files and create DataFrame objects.
You’ve used the Pandas read_csv() and .to_csv() methods to read and write CSV files. You also used similar methods to read and write Excel, JSON, HTML, SQL, and pickle files. These functions are very convenient and widely used. They allow you to save or load your data in a single function or method call.
You’ve also learned how to save time, memory, and disk space when working with large data files:
- Compress or decompress files
- Choose the rows and columns you want to load
- Use less precise data types
- Split data into chunks and process them one by one
You’ve mastered a significant step in the machine learning and data science process! If you have any questions or comments, then please put them in the comments section below.
[ Improve Your Python With 🐍 Python Tricks 💌 – Get a short & sweet Python Trick delivered to your inbox every couple of days. >> Click here to learn more and see examples ]

 Been a long ten years, but it's flown past. This particular decade
happens to coincide with my first years of full-time professional
software engineering.
Been a long ten years, but it's flown past. This particular decade
happens to coincide with my first years of full-time professional
software engineering.

 Problem of Too Many Widgets.png
Problem of Too Many Widgets.png
 Problem of Too Many Widgets With Fixed Heights
Problem of Too Many Widgets With Fixed Heights
 Qt Creator — Select MainWindow for widget type
Qt Creator — Select MainWindow for widget type
 Add Scroll Area
Add Scroll Area
 Lay Out The Scroll Area Vertically Or Horizontally
Lay Out The Scroll Area Vertically Or Horizontally
 The Scroll Area Is Created
The Scroll Area Is Created
 Add Labels to The Scroll Area And Set the Layout
Add Labels to The Scroll Area And Set the Layout
 Adding More Labels to QScrollArea
Adding More Labels to QScrollArea
 Setting Fixed Heights for Labels
Setting Fixed Heights for Labels
 ScrollArea Properties
ScrollArea Properties
 App With Scroll Bar
App With Scroll Bar
 Scroll Area App
Scroll Area App