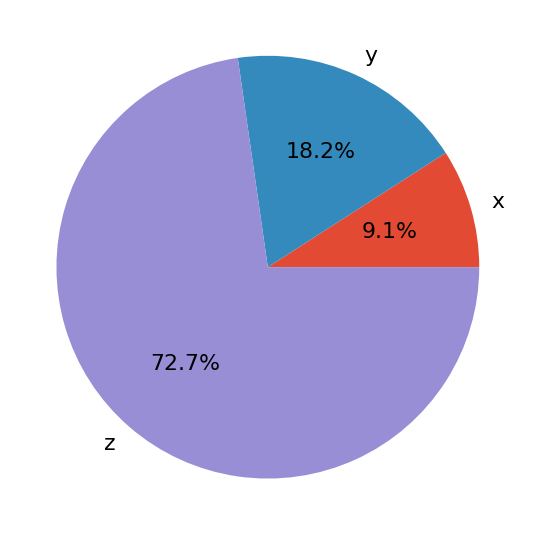
Python Bytes: #160 Your JSON shall be streamed
Steve Dower: What makes Python a great language?
I know I’m far from the only person who has opined about this topic, but figured I’d take my turn.
A while ago I hinted on Twitter that I have Thoughts(tm) about the future of Python, and while this is not going to be that post, this is going to be important background for when I do share those thoughts.
If you came expecting a well researched article full of citations to peer-reviewed literature, you came to the wrong place. Similarly if you were hoping for unbiased and objective analysis. I’m not even going to link to external sources for definitions. This is literally just me on a soap box, and you can take it or leave it.
I’m also deliberately not talking about CPython the runtime, pip the package manager, venv the %PATH% manipulator, or PyPI the ecosystem. This post is about the Python language.
My hope is that you will get some ideas for thinking about why some programming languages feel better than others, even if you don’t agree that Python feels better than most.
Need To Know
What makes Python a great language? It gets the need to know balance right.
When I use the term “need to know”, I think of how the military uses the term. For many, “need to know” evokes thoughts of power imbalances, secrecy, and dominance-for-the-sake-of-dominance. But even in cases that may look like or actually be as bad as these, the intent is to achieve focus.
In a military organisation, every individual needs to make frequent life-or-death choices. The more time you spend making each choice, the more likely you are choosing death (specifically, your own). Having to factor in the full range of ethical factors into every decision is very inefficient.
Since no army wants to lose their own men, they delegate decision-making up through a series of ranks. By the time individuals are in the field, the biggest decisions are already made, and the soldier has a very narrow scope to make their own decisions. They can focus on exactly what they need to know, trusting that their superiors have taken into account anything else that they don’t need to know.
Software libraries and abstractions are fundamentally the same. Another developer has taken the broader context into account, and has provided you – the end-developer – with only what you need to know. You get to focus on your work, trusting that the rest has been taken care of.
Memory management is probably the easiest example. Languages that decide how memory management is going to work (such as through a garbage collector) have taken that decision for you. You don’t need to know. You get to use the time you would have been thinking about deallocation to focus on your actual task.
Does “need to know” ever fail? Of course it does. Sometimes you need more context in order to make a good decision. In a military organisation, there are conventions for requesting more information, ways to get promoted into positions with more context (and more complex decisions), and systems for refusing to follow orders (which mostly don’t turn out so well for the person refusing, but hey, there’s a system).
In software, “need to know” breaks down when you need some functionality that isn’t explicitly exposed or documented, when you need to debug library or runtime code, or just deal with something not behaving as it claims it should. When these situations arise, not being able to incrementally increase what you know becomes a serious blockage.
A good balance of “need to know” will actively help you focus on getting your job done, while also providing the escape hatches necessary to handle the times you need to know more. Python gets this balance right.
Python’s Need To Know levels
There are many levels of what you “need to know” to use Python.
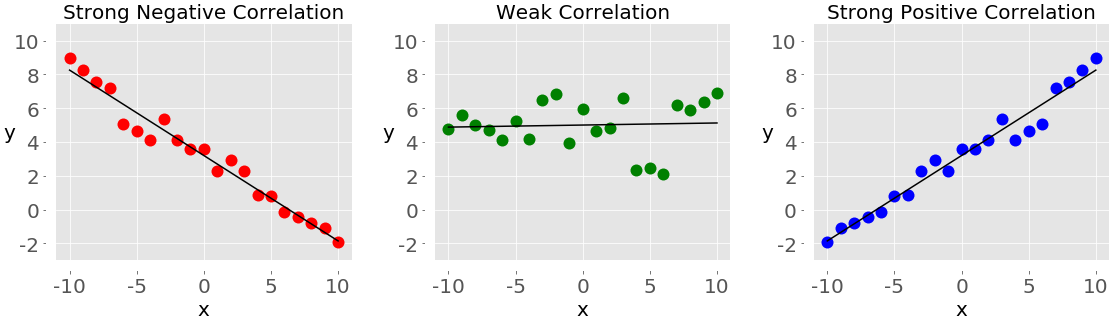
At the lowest level, there’s the basic syntax and most trivial semantics of assignment, attributes and function calls. These concepts, along with your project-specific context, are totally sufficient to write highly effective code.
The example to the right (source) generates a histogram from a random distribution. By my count, there are two distinct words in that are not specific to the task at hand (“import” and “as”), and the places they are used are essentially boiler-plate – they were likely copied by the author, rather than created by the author. Everything else in the sample code relates to specifying the random distribution and creating the plot.
The most complex technical concept used is tuple unpacking, but all the user needs to know here is that they’re getting multiple return values. The fact that there’s really only a single return value and that the unpacking is performed by the assignment isn’t necessary or useful knowledge.
Find a friend who’s not a developer and try this experiment on them: show them
x, y = get_points()and explain how it works, without ever mentioning that it’s returning multiple values. Then point out thatget_points()actually just returns two values, andx, y =is how you give them names. Turns out, they won’t need to know how it works, just what it does.
As you add introduce new functionality, you will see the same pattern repeated. for x in y: can (and should) be explained without mentioning iterators. open() can (and should) be explained without mentioning the io module. Class instantiation can (and should) be explained without mentioning __call__. And so on.
Python very effectively hides unnecessary details from those who just want to use it.
Think about basically any other language you’ve used. How many concepts do you need to express the example above?
Basically every other language is going to distinguish between declaring a variable and assigning a variable. Many are going to require nominal typing, where you need to know about types before you can do assignment. I can’t think of many languages with fewer than the three concepts Python requires to generate a histogram from a random distribution with certain parameters (while also being readable from top to bottom – yes, I thought of LISP).
When Need To Know breaks down
But when need to know starts breaking down, Python has some of the best escape hatches in the entire software industry.
For starters, there are no truly private members. All the code you use in your Python program belongs to you. You can read everything, mutate everything, wrap everything, proxy everything, and nobody can stop you. Because it’s your program. Duck typing makes a heroic appearance here, enabling new ways to overcome limiting abstractions that would be fundamentally impossible in other languages.
Should you make a habit of doing this? Of course not. You’re using libraries for a reason – to help you focus on your own code by delegating “need to know” decisions to someone else. If you are going to regularly question and ignore their decisions, you completely spoil any advantage you may have received. But Python also allows you to rely on someone else’s code without becoming a hostage to their choices.
Today, the Python ecosystem is almost entirely publicly-visible code. You don’t need to know how it works, but you have the option to find out. And you can find out by following the same patterns that you’re familiar with, rather than having to learn completely new skills. Reading Python code, or interactively inspecting live object graphs, are exactly what you were doing with your own code.
Compare Python to languages that tend towards sharing compiled, minified, packaged or obfuscated code, and you’ll have a very different experience figuring out how things really (don’t) work.
Compare Python to languages that emphasize privacy, information hiding, encapsulation and nominal typing, and you’ll have a very different experience overcoming a broken or limiting abstraction.
Features you don’t Need To Know about
In the earlier plot example, you didn’t need to know about anything beyond assignment, attributes and function calls. How much more do you need to know to use Python? And who needs to know about these extra features?
As it turns out, there are millions of Python developers who don’t need much more than assignment, attributes and function calls. Those of us in the 1% of the Python community who use Twitter and mailing lists like to talk endlessly about incredibly advanced features, such as assignment expressions and position-only parameters, but the reality is that most Python users never need these and should never have to care.
When I teach introductory Python programming, my order of topics is roughly assignment, arithmetic, function calls (with imports thrown in to get to the interesting ones), built-in collection types, for loops, if statements, exception handling, and maybe some simple function definitions and decorators to wrap up. That should be enough for 90% of Python careers (syntactically – learning which functions to call and when is considerably more effort than learning the language).
The next level up is where things get interesting. Given the baseline knowledge above, the Python’s next level allows 10% of developers to provide the 90% with significantly more functionality without changing what they need to know about the language. Those awesome libraries are written by people with deeper technical knowledge, but (can/should) expose only the simplest syntactic elements.
When I adopt classes, operator overloading, generators, custom collection types, type checking, and more, Python does not force my users to adopt them as well. When I expand my focus to include more complexity, I get to make decisions that preserve my users’ need to know.
For example, my users know that calling something returns a value, and that returned values have attributes or methods. Whether the callable is a function or a class is irrelevant to them in Python. But compare with most other languages, where they would have to change their syntax if I changed a function into a class.
When I change a function to return a custom mapping type rather than a standard dictionary, it is irrelevant to them. In other languages, the return type is also specified explicitly in my user’s code, and so even a compatible change might force them outside of what they really need to know.
If I return a number-like object rather than a built-in integer, my users don’t need to know. Most languages don’t have any way to replace primitive types, but Python provides all the functionality I need to create a truly number-like object.
Clearly the complexity ramps up quickly, even at this level. But unlike most other languages, complexity does not travel down. Just because some complexity is used within your codebase doesn’t mean you will be forced into using it everywhere throughout the codebase.
The next level adds even more complexity, but its use also remains hidden behind normal syntax. Metaclasses, object factories, decorator implementations, slots, __getattribute__ and more allow a developer to fundamentally rewrite how the language works. There’s maybe 1% of Python developers who ever need to be aware of these features, and fewer still who should use them, but the enabling power is unique among languages that also have such an approachable lowest level.
Even with this ridiculous level of customisation, the same need to know principles apply, and in a way that only Python can do it. Enums and data classes in Python are based on these features, but the knowledge required to use them is not the same as the knowledge required to create them. Users get to focus on what they’re doing, assisted by trusting someone else to have made the right decision about what they need to know.
Summary and foreshadowing
People often cite Python’s ecosystem as the main reason for its popularity. Others claim the language’s simplicity or expressiveness is the primary reason.
I would argue that the Python language has an incredibly well-balanced sense of what developers need to know. Better than any other language I’ve used.
Most developers get to write incredibly functional and focused code with just a few syntax constructs. Some developers produce reusable functionality that is accessible through simple syntax. A few developers manage incredible complexity to provide powerful new semantics without leaving the language.
By actively helping library developers write complex code that is not complex to use, Python has been able to build an amazing ecosystem. And that amazing ecosystem is driving the popularity of the language.
But does our ecosystem have the longevity to maintain the language…? Does the Python language have the qualities to survive a changing ecosystem…? Will popular libraries continue to drive the popularity of the language, or does something need to change…?
(Contact me on Twitter for discussion.)
Kushal Das: Updates on Unoon in December 2019
This Saturday evening, I sat with Unoon project after a few weeks, I was continuously running it, but, did not resume the development effort. This time Bhavin also joined me. Together, we fixed a location of the whitelist files issue, and unoon now also has a database (using SQLite), which stores all the historical process and connection information. In the future, we will provide some way to query this information.
As usual, we learned many new things about different Linux processes while doing this development. One of the important ones is about running podman process, and how the user id maps to the real system. Bhavin added a patch that fixes a previously known issue of crashing due to missing user name. Now, unoon shows the real user ID when it can not find the username in the /etc/passwd file.
You can read about Unoon more in my previous blog post.
Kushal Das: Highest used usernames in break-in attempts to my servers 2019

A few days ago, I wrote about different IP addresses trying to break into my servers. Today, I looked into another server to find the frequently used user names used in the SSH attempts.
- admin 36228
- test 19249
- user 17164
- ubuntu 16233
- postgres 16217
- oracle 9738
- git 8118
- ftpuser 7028
- teamspea 6560
- mysql 5650
- nagios 5599
- pi 5239
- deploy 5167
- hadoop 5011
- guest 4798
- dev 4468
- ts3 4277
- minecraf 4145
- support 3940
- ubnt 3549
- debian 3515
- demo 3489
- tomcat 3435
- vagrant 3042
- zabbix 3033
- jenkins 3027
- develope 2941
- sinusbot 2914
- user1 2898
- administ 2747
- bot 2590
- testuser 2459
- ts 2403
- apache 2391
- www 2329
- default 2293
- odoo 2168
- test2 2161
- backup 2133
- steam 2129
- 1234 2026
- server 1890
- www-data 1853
- web 1850
- centos 1796
- vnc 1783
- csgoserv 1715
- prueba 1677
- test1 1648
- a 1581
- student 1568
- csgo 1524
- weblogic 1522
- ts3bot 1521
- mc 1434
- gpadmin 1427
- redhat 1378
- alex 1375
- system 1362
- manager 1359
I never knew that admin is such important user name for Linux servers, I thought I will see root there. Also, why alex? I can under the reason behind pi. If you want to find out the similar details, you can use the following command.
last -f /var/log/btmp
Programiz: Python Dictionary Comprehension
Steve Dower: What makes Python a great language?
I know I’m far from the only person who has opined about this topic, but figured I’d take my turn.
A while ago I hinted on Twitter that I have Thoughts(tm) about the future of Python, and while this is not going to be that post, this is going to be important background for when I do share those thoughts.
If you came expecting a well researched article full of citations to peer-reviewed literature, you came to the wrong place. Similarly if you were hoping for unbiased and objective analysis. I’m not even going to link to external sources for definitions. This is literally just me on a soap box, and you can take it or leave it.
I’m also deliberately not talking about CPython the runtime, pip the package manager, venv the %PATH% manipulator, or PyPI the ecosystem. This post is about the Python language.
My hope is that you will get some ideas for thinking about why some programming languages feel better than others, even if you don’t agree that Python feels better than most.
Need To Know
What makes Python a great language? It gets the need to know balance right.
When I use the term “need to know”, I think of how the military uses the term. For many, “need to know” evokes thoughts of power imbalances, secrecy, and dominance-for-the-sake-of-dominance. But even in cases that may look like or actually be as bad as these, the intent is to achieve focus.
In a military organisation, every individual needs to make frequent life-or-death choices. The more time you spend making each choice, the more likely you are choosing death (specifically, your own). Having to factor in the full range of ethical factors into every decision is very inefficient.
Since no army wants to lose their own men, they delegate decision-making up through a series of ranks. By the time individuals are in the field, the biggest decisions are already made, and the soldier has a very narrow scope to make their own decisions. They can focus on exactly what they need to know, trusting that their superiors have taken into account anything else that they don’t need to know.
Software libraries and abstractions are fundamentally the same. Another developer has taken the broader context into account, and has provided you – the end-developer – with only what you need to know. You get to focus on your work, trusting that the rest has been taken care of.
Memory management is probably the easiest example. Languages that decide how memory management is going to work (such as through a garbage collector) have taken that decision for you. You don’t need to know. You get to use the time you would have been thinking about deallocation to focus on your actual task.
Does “need to know” ever fail? Of course it does. Sometimes you need more context in order to make a good decision. In a military organisation, there are conventions for requesting more information, ways to get promoted into positions with more context (and more complex decisions), and systems for refusing to follow orders (which mostly don’t turn out so well for the person refusing, but hey, there’s a system).
In software, “need to know” breaks down when you need some functionality that isn’t explicitly exposed or documented, when you need to debug library or runtime code, or just deal with something not behaving as it claims it should. When these situations arise, not being able to incrementally increase what you know becomes a serious blockage.
A good balance of “need to know” will actively help you focus on getting your job done, while also providing the escape hatches necessary to handle the times you need to know more. Python gets this balance right.
Python’s Need To Know levels
There are many levels of what you “need to know” to use Python.
At the lowest level, there’s the basic syntax and most trivial semantics of assignment, attributes and function calls. These concepts, along with your project-specific context, are totally sufficient to write highly effective code.
The example to the right (source) generates a histogram from a random distribution. By my count, there are two distinct words in that are not specific to the task at hand (“import” and “as”), and the places they are used are essentially boiler-plate – they were likely copied by the author, rather than created by the author. Everything else in the sample code relates to specifying the random distribution and creating the plot.
The most complex technical concept used is tuple unpacking, but all the user needs to know here is that they’re getting multiple return values. The fact that there’s really only a single return value and that the unpacking is performed by the assignment isn’t necessary or useful knowledge.
Find a friend who’s not a developer and try this experiment on them: show them
x, y = get_points()and explain how it works, without ever mentioning that it’s returning multiple values. Then point out thatget_points()actually just returns two values, andx, y =is how you give them names. Turns out, they won’t need to know how it works, just what it does.
As you add introduce new functionality, you will see the same pattern repeated. for x in y: can (and should) be explained without mentioning iterators. open() can (and should) be explained without mentioning the io module. Class instantiation can (and should) be explained without mentioning __call__. And so on.
Python very effectively hides unnecessary details from those who just want to use it.
Think about basically any other language you’ve used. How many concepts do you need to express the example above?
Basically every other language is going to distinguish between declaring a variable and assigning a variable. Many are going to require nominal typing, where you need to know about types before you can do assignment. I can’t think of many languages with fewer than the three concepts Python requires to generate a histogram from a random distribution with certain parameters (while also being readable from top to bottom – yes, I thought of LISP).
When Need To Know breaks down
But when need to know starts breaking down, Python has some of the best escape hatches in the entire software industry.
For starters, there are no truly private members. All the code you use in your Python program belongs to you. You can read everything, mutate everything, wrap everything, proxy everything, and nobody can stop you. Because it’s your program. Duck typing makes a heroic appearance here, enabling new ways to overcome limiting abstractions that would be fundamentally impossible in other languages.
Should you make a habit of doing this? Of course not. You’re using libraries for a reason – to help you focus on your own code by delegating “need to know” decisions to someone else. If you are going to regularly question and ignore their decisions, you completely spoil any advantage you may have received. But Python also allows you to rely on someone else’s code without becoming a hostage to their choices.
Today, the Python ecosystem is almost entirely publicly-visible code. You don’t need to know how it works, but you have the option to find out. And you can find out by following the same patterns that you’re familiar with, rather than having to learn completely new skills. Reading Python code, or interactively inspecting live object graphs, are exactly what you were doing with your own code.
Compare Python to languages that tend towards sharing compiled, minified, packaged or obfuscated code, and you’ll have a very different experience figuring out how things really (don’t) work.
Compare Python to languages that emphasize privacy, information hiding, encapsulation and nominal typing, and you’ll have a very different experience overcoming a broken or limiting abstraction.
Features you don’t Need To Know about
In the earlier plot example, you didn’t need to know about anything beyond assignment, attributes and function calls. How much more do you need to know to use Python? And who needs to know about these extra features?
As it turns out, there are millions of Python developers who don’t need much more than assignment, attributes and function calls. Those of us in the 1% of the Python community who use Twitter and mailing lists like to talk endlessly about incredibly advanced features, such as assignment expressions and position-only parameters, but the reality is that most Python users never need these and should never have to care.
When I teach introductory Python programming, my order of topics is roughly assignment, arithmetic, function calls (with imports thrown in to get to the interesting ones), built-in collection types, for loops, if statements, exception handling, and maybe some simple function definitions and decorators to wrap up. That should be enough for 90% of Python careers (syntactically – learning which functions to call and when is considerably more effort than learning the language).
The next level up is where things get interesting. Given the baseline knowledge above, the Python’s next level allows 10% of developers to provide the 90% with significantly more functionality without changing what they need to know about the language. Those awesome libraries are written by people with deeper technical knowledge, but (can/should) expose only the simplest syntactic elements.
When I adopt classes, operator overloading, generators, custom collection types, type checking, and more, Python does not force my users to adopt them as well. When I expand my focus to include more complexity, I get to make decisions that preserve my users’ need to know.
For example, my users know that calling something returns a value, and that returned values have attributes or methods. Whether the callable is a function or a class is irrelevant to them in Python. But compare with most other languages, where they would have to change their syntax if I changed a function into a class.
When I change a function to return a custom mapping type rather than a standard dictionary, it is irrelevant to them. In other languages, the return type is also specified explicitly in my user’s code, and so even a compatible change might force them outside of what they really need to know.
If I return a number-like object rather than a built-in integer, my users don’t need to know. Most languages don’t have any way to replace primitive types, but Python provides all the functionality I need to create a truly number-like object.
Clearly the complexity ramps up quickly, even at this level. But unlike most other languages, complexity does not travel down. Just because some complexity is used within your codebase doesn’t mean you will be forced into using it everywhere throughout the codebase.
The next level adds even more complexity, but its use also remains hidden behind normal syntax. Metaclasses, object factories, decorator implementations, slots, __getattribute__ and more allow a developer to fundamentally rewrite how the language works. There’s maybe 1% of Python developers who ever need to be aware of these features, and fewer still who should use them, but the enabling power is unique among languages that also have such an approachable lowest level.
Even with this ridiculous level of customisation, the same need to know principles apply, and in a way that only Python can do it. Enums and data classes in Python are based on these features, but the knowledge required to use them is not the same as the knowledge required to create them. Users get to focus on what they’re doing, assisted by trusting someone else to have made the right decision about what they need to know.
Summary and foreshadowing
People often cite Python’s ecosystem as the main reason for its popularity. Others claim the language’s simplicity or expressiveness is the primary reason.
I would argue that the Python language has an incredibly well-balanced sense of what developers need to know. Better than any other language I’ve used.
Most developers get to write incredibly functional and focused code with just a few syntax constructs. Some developers produce reusable functionality that is accessible through simple syntax. A few developers manage incredible complexity to provide powerful new semantics without leaving the language.
By actively helping library developers write complex code that is not complex to use, Python has been able to build an amazing ecosystem. And that amazing ecosystem is driving the popularity of the language.
But does our ecosystem have the longevity to maintain the language…? Does the Python language have the qualities to survive a changing ecosystem…? Will popular libraries continue to drive the popularity of the language, or does something need to change…?
(Contact me on Twitter for discussion.)
Peter Bengtsson: A Python and Preact app deployed on Heroku
Heroku is great but it's sometimes painful when your app isn't just in one single language. What I have is a project where the backend is Python (Django) and the frontend is JavaScript (Preact). The folder structure looks like this:
/
- README.md
- manage.py
- requirements.txt
- my_django_app/
- settings.py
- asgi.py
- api/
- urls.py
- views.py
- frontend/
- package.json
- yarn.lock
- preact.config.js
- build/
...
- src/
...A bunch of things omitted for brevity but people familiar with Django and preact-cli/create-create-app should be familiar.
The point is that the root is a Python app and the front-end is exclusively inside a sub folder.
When you do local development, you start two servers:
./manage.py runserver- startshttp://localhost:8000cd frontend && yarn start- startshttp://localhost:3000
The latter is what you open in your browser. That preact app will do things like:
constresponse=awaitfetch('/api/search');and, in preact.config.js I have this:
exportdefault(config,env,helpers)=>{if(config.devServer){config.devServer.proxy=[{path:"/api/**",target:"http://localhost:8000"}];}};...which is hopefully self-explanatory. So, calls like GET http://localhost:3000/api/search actually goes to http://localhost:8000/api/search.
That's when doing development. The interesting thing is going into production.
Before we get into Heroku, let's first "merge" the two systems into one and the trick used is Whitenoise. Basically, Django's web server will be responsibly not only for things like /api/search but also static assets such as / --> frontend/build/index.html and /bundle.17ae4.js --> frontend/build/bundle.17ae4.js.
This is basically all you need in settings.py to make that happen:
MIDDLEWARE=["django.middleware.security.SecurityMiddleware","whitenoise.middleware.WhiteNoiseMiddleware",...]WHITENOISE_INDEX_FILE=TrueSTATIC_URL="/"STATIC_ROOT=BASE_DIR/"frontend"/"build"However, this isn't quite enough because the preact app uses preact-router which uses pushState() and other code-splitting magic so you might have a URL, that users see, like this: https://myapp.example.com/that/thing/special and there's nothing about that in any of the Django urls.py files. Nor is there any file called frontend/build/that/thing/special/index.html or something like that.
So for URLs like that, we have to take a gamble on the Django side and basically hope that the preact-router config knows how to deal with it. So, to make that happen with Whitenoise we need to write a custom middleware that looks like this:
fromwhitenoise.middlewareimportWhiteNoiseMiddlewareclassCustomWhiteNoiseMiddleware(WhiteNoiseMiddleware):defprocess_request(self,request):ifself.autorefresh:static_file=self.find_file(request.path_info)else:static_file=self.files.get(request.path_info)# These two lines is the magic.# Basically, the URL didn't lead to a file (e.g. `/manifest.json`)# it's either a API path or it's a custom browser path that only# makes sense within preact-router. If that's the case, we just don't# know but we'll give the client-side preact-router code the benefit# of the doubt and let it through.ifnotstatic_fileandnotrequest.path_info.startswith("/api"):static_file=self.files.get("/")ifstatic_fileisnotNone:returnself.serve(static_file,request)And in settings.py this change:
MIDDLEWARE = [
"django.middleware.security.SecurityMiddleware",
- "whitenoise.middleware.WhiteNoiseMiddleware",+ "my_django_app.middleware.CustomWhiteNoiseMiddleware",
...
]
Now, all traffic goes through Django. Regular Django view functions, static assets, and everything else fall back to frontend/build/index.html.
Heroku
Heroku tries to make everything so simple for you. You basically, create the app (via the cli or the Heroku web app) and when you're ready you just do git push heroku master. However that won't be enough because there's more to this than Python.
Unfortunately, I didn't take notes of my hair-pulling excruciating journey of trying to add buildpacks and hacks and Procfiles and custom buildpacks. Nothing seemed to work. Perhaps the answer was somewhere in this issue: "Support running an app from a subdirectory" but I just couldn't figure it out. I still find buildpacks confusing when it's beyond Hello World. Also, I didn't want to run Node as a service, I just wanted it as part of the "build process".
Docker to the rescue
Finally I get a chance to try "Deploying with Docker" in Heroku which is a relatively new feature. And the only thing that scared me was that now I need to write a heroku.yml file which was confusing because all I had was a Dockerfile. We'll get back to that in a minute!
So here's how I made a Dockerfile that mixes Python and Node:
FROMnode:12asfrontendCOPY . /app
WORKDIR /appRUNcd frontend && yarn install && yarn build
FROMpython:3.8-slimWORKDIR /appRUN groupadd --gid 10001 app && useradd -g app --uid 10001 --shell /usr/sbin/nologin app
RUN chown app:app /tmp
RUN apt-get update &&\
apt-get upgrade -y &&\
apt-get install -y --no-install-recommends \
gcc apt-transport-https python-dev
# Gotta try moving this to poetry instead!COPY ./requirements.txt /app/requirements.txt
RUN pip install --upgrade --no-cache-dir -r requirements.txt
COPY . /app
COPY --from=frontend /app/frontend/build /app/frontend/build
USER appENVPORT=8000EXPOSE $PORTCMD uvicorn gitbusy.asgi:application --host 0.0.0.0 --port $PORTIf you're not familiar with it, the critical trick is on the first line where it builds some Node with as frontend. That gives me a thing I can then copy from into the Python image with COPY --from=frontend /app/frontend/build /app/frontend/build.
Now, at the very end, it starts a uvicorn server with all the static .js, index.html, and favicon.ico etc. available to uvicorn which ultimately runs whitenoise.
To run and build:
docker build . -t my_app docker run -t -i --rm --env-file .env -p 8000:8000 my_app
Now, opening http://localhost:8000/ is a production grade app that mixes Python (runtime) and JavaScript (static).
Heroku + Docker
Heroku says to create a heroku.yml file and that makes sense but what didn't make sense is why I would add cmd line in there when it's already in the Dockerfile. The solution is simple: omit it. Here's what my final heroku.yml file looks like:
build:docker:web:DockerfileCheck in the heroku.yml file and git push heroku master and voila, it works!
To see a complete demo of all of this check out https://github.com/peterbe/gitbusy and https://gitbusy.herokuapp.com/
Weekly Python StackOverflow Report: (ccvi) stackoverflow python report
Between brackets: [question score / answers count]
Build date: 2019-12-14 12:53:35 GMT
- Python __getitem__ and in operator result in strange behavior - [22/1]
- What is the reason for difference between integer division and float to int conversion in python? - [18/1]
- Match multiple(3+) occurrences of each character - [7/5]
- Pandas Dataframe: Multiplying Two Columns - [7/3]
- What does !r mean in Python? - [7/1]
- python how to find the number of days in each month from Dec 2019 and forward between two date columns - [6/4]
- Itertools zip_longest with first item of each sub-list as padding values in stead of None by default - [6/4]
- Writing more than 50 millions from Pyspark df to PostgresSQL, best efficient approach - [6/0]
- How to merge and groupby between seperate dataframes - [5/3]
- Setting the index after merging with pandas? - [5/3]
Catalin George Festila: Python 3.7.5 : Django admin shell by Grzegorz Tężycki.
Andre Roberge: A Tiny Python Exception Oddity
- The inconsistent behaviour is so tiny, that I doubt most people would notice - including myself before working on Friendly-traceback.
- This is SyntaxError that is not picked up by flake8; however, pylint does pick it up.
- By Python, I mean CPython. After trying to figure out why this case was different, I downloaded Pypy and saw that Pypy did not show the odd behaviour.
- To understand the origin of this different behaviour, one needs to look at some obscure inner parts of the CPython interpreter.
- This would likely going to be found totally irrelevant by 99.999% of Python programmers. If you are not the type of person who is annoyed by tiny oddities, you probably do not want to read any further.
Normal behaviour
Odd case
Why does it matter to me?
Why is this case handled differently by CPython?
Conclusion
Anwesha Das: Rootconf Hyderbad, 2019
What is Rootconf?
Rootconf is the conference on sysadmins, DevOps, SRE, Network engineers. Rootconf started its journey in 2012 in Bangalore, 2019 was the 7th edition of Rootconf. In these years, through all the Rootconfs, there is a community that has developed around Rootconf. Now people do come to attend Rootconf not just to attend the conference but also to attend friends and peers to discuss projects and ideas.
Need for more Rootconf
Over all these years, we have witnessed changes in the network, infrastructure, and security threats. We have designed Rootconf (in all these years), keeping in mind the changing needs of the community. Lately, we have realized that the needs of the community based on their geographic locations/ cities. Like in Pune, there is a considerable demand for sessions that deals with small size infrastructure suited for startups and SMEs as there is a growing startup industry there. In Delhi, there is a demand for discussion around data centers, network designs, and so on. And in Hyderabad, there is a want for solutions around large scale infrastructure. The Bangalore event did not suffice to solve all these needs. So more the merrier, we decided to have more than one Rootconf a year.
Rootconf Pune was the first of this 'outstation Rootconf journey'. The next was Rootconf Hyderabad. It was the first event for which I was organizing the editorial, community, and all by myself.
I joined HasGeek as Community Manager and Editorial co-ordinator. After my Rootconf, Bangalore Zainab fixed a goal for me.
Z : 'Anwesha, I want to organize Rootconf Hyderabad all by yourself, you must be doing with no or minimum help from me.'
A: "Ummm hmmm ooops"
Z: 'Do not worry, I will be there to guide you. We will have our test run with you in Pune. So buck up, girl.'
Rootconf Hyderabad, the conference
The preparation for Rootconf Hyderabad started with them. After months of the editorial process - scouting for the proposals, reviewing them, having several rehearsals, and after passing the iron test in Pune, I reached Hyderabad to join my colleague Mak. Mak runs the sales at Hasgeek. With the camera, we had our excellent AV captain Amogh. So I was utterly secured and not worried about those two aspects.
A day before the conference Damini, our emcee, and I chalked out the plans for navigating the schedule and coordinating the conference. We met the volunteers at the venue after a humongous lunch with Hyderabadi deliciously (honest confession: food is the primary reason why I love to attend the conference in Hyderabad). We have several call volunteers in which our volunteer coordinator Jyotsna briefed them the duties. But it is always essential to make the volunteers introduced with the ground reality. We had a meet up at Thought Works.
The day of the conference starts early, much too early for the organizers and volunteers. Rootconf Hyderabad was no different. We opened the registration, and people started flocking in the auditorium. I opened the conference by addressing -
- What is Rootconf?
- The journey of Rootconf.
- Why we need several editions of Rootconf in different geographical locations all across India?
Then our emcee Damini took over. The first half of our schedule designed keeping the problems of large scale infrastructure in mind, like observability, maintainability, scalability, performance, taming the large systems, and networking issues. Piyush started our first speaker gave a talk on Observability and control theory. Next was Flipkart's journey of "Fast object distribution using P2P" by Ankur Jain. After a quick beverage break, Anubhav Mishra shared his take on "Taming infrastructure workflow at scale", the story of Hashicorp Followed by Tasdik Rahman and his story of "Achieving repeatable, extensible and self serve infrastructure" at Gojek."
The next half of the day planned to address the issues shared with infrastructure despite size or complexity. Like - Security, DevOpSec, scaling, and of course, microservices (an infrastructure conference seems incomplete without the discussion around monolith to microservices). Our very own security expert Lavakumar started it with "Deploying and managing CSP: the browser-side firewall", describing the security complexities post mege cart attack days. Jambunathan shared the tale of "Designing microservices around your data design” . For the last talk of the day, we had Gaurav Kamboj. He told us what happens with the system engineers at Hotstar when Virat Kohli is batting on his 90s, in "Scaling hotstar.com for 25 million concurrent viewers"
Birds of a Feather (BOF) session has always been a favorite at Rootconf. These non-recorded sessions give the participants a chance, to be frank. We have facilitators to progress the discussion and not presenters. While we had talks going on in the main audi, there are dedicated BOF area where we had sessions on
- AI Ops facilitated by Jayesh Bapu Ahire and Jambunathan Valady,
- Infrastructure as Code facilitated by Anubhav Mishra, Tasdik Rahman
- Observability by Gaurav.
This was the first time gauging the popularity of the BOFs we tried something new. We had a BOF session planned at the primary audi. It was on "Doing DevSecOps in your organization," aided by Lava and Hari. It was one session in which our emcee Damini had a difficult time to end. People had so many stories to share questions to ask, but there was no time. I also got some angry looks (which I do not mind at all) :).
In India, I have noticed that most of the conferences fail to have good/up to the mark flash talks. Invariably they have community information, conference, or meetup notifications (the writer is guilty of doing it). So I proposed that why can not we accept proposals for flash talks as well. Half of them are pre-selected and rest selected on the spot. Zainab agreed to it. Now we are following this rule since Rootconf Pune, and the quality of the flash talks has improved a lot. We had some fantastic flash talks. You can check it for yourself at https://www.youtube.com/watch?v=AlREWUAEMVk.
Thank you
Organizing a conference is not a person's job. In an extensive infrastructure, it is the small tools, microservices that keeps an extensive system working. Consider conference as a system, tasks as microservices. It requires each task to be perfect for the conference to be successful and flawless. And I am blessed to have an amazing team. Each amazing volunteers, the Null Hyderabad, Mozilla, AWS community, our emcee Damini, Hall manager Geetanjali, Speakers, Sponsors, attendees, and my team HasGeek. Last but not least, thank you, Zainab, for trusting me, being by my side, and not letting me fall.
The experience
Organizing a conference has been the journey of estrogen and adrenaline overflow for me. Be it getting into nightmares the excitement of each ticket sales, the long chats with the reviewers about talks, BOFs, discussion with the communities what they want from Rootconf, jitters before the conference starts or tweets, a blog post from the people that they enjoyed the conference was useful for them. It was an exciting, scary, happy, and satisfying journey for me. And guess what, my life continues to be so as Rootconf is ready with it's Delhi edition. I hope to meet you there.
Catalin George Festila: Python 3.7.5 : Simple intro in CSRF.
S. Lott: Functional programming design pattern: Nested Iterators == Flattening
Consider this data collection process.
for h in some_high_level_collection(arg1):
for l in h.some_low_level_collection(arg2):
if some_filter(l):
logger.info("Processing %s %s", h, l)
some_function(h, l)
Here's something that's a little easier to work with:
def h_l_iter(arg1, arg2):
for h in some_high_level_collection(arg1):
for l in h.some_low_level_collection(arg2):
if some_filter(l):
logger.info("Processing %s %s", h, l)
yield h, l
itertools.starmap(some_function, h_l_iter(arg1, arg2))
I think the pattern here is a kind of "Flattened Map" transformation. The initial design, with nested loops wrapping a process wasn't a good plan. A better plan is to think of the nested loops as a way to flatten the two tiers of the hierarchy into a single iterator. Then a mapping can be applied to process each item from that flat iterator.
Extracting the Filter
We can now tease apart the nested loops to expose the filter. In the version above, the body of the h_l_iter() function binds log-writing with the yield. If we take those two apart, we gain the flexibility of being able to change the filter (or the logging) without an awfully complex rewrite.T = TypeVar('T')
def logging_iter(source: Iterable[T]) -> Iterator[T]:
for item in source:
logger.info("Processing %s", item)
yield item
def h_l_iter(arg1, arg2):
for h in some_high_level_collection(arg1):
for l in h.some_low_level_collection(arg2):
yield h, l
raw_data = h_l_iter(arg1, arg2)
filtered_subset = logging_iter(filter(some_filter, raw_data))
itertools.starmap(some_function, filtered_subset)
Now, I can introduce various forms of multiprocessing to improve concurrency.
This transformed a hard-wired set of nest loops, if, and function evaluation into a "Flattener" that can be combined with off-the shelf filtering and mapping functions.
I've snuck in a kind of "tee" operation that writes an iterable sequence to a log. This can be injected at any point in the processing.
Logging the entire "item" value isn't really a great idea. Another mapping is required to create sensible log messages from each item. I've left that out to keep this exposition more focused.
Full Flattening
The h_l_iter() function is actually a generator expression. A function isn't needed.h_l_iter = (
(h, l)
for h in some_high_level_collection(arg1)
for l in h.some_low_level_collection(arg2)
)
This simplification doesn't add much value, but it seems to be general truth. In Python, it's a small change in syntax and therefore, an easy optimization to make.
What About The File System?
def recursive_walk(path: Path) -> Iterator[Path]:
for item in path.glob():
if item.is_file():
yield item
elif item.is_dir():
yield from recursive_walk(item)
tl;dr
The too-long version of this is:This pattern works for simple, flat items, nested structures, and even recursively-defined trees. It introduces flexibility with no real cost.
The other pattern in play is:
These felt like blinding revelations to me.
Codementor: Function-Based Views vs Class-Based Views in Django
Wesley Chun: Authorized Google API access from Python (part 2 of 2)
NOTE: You can also watch a video walkthrough of the common code covered in this blogpost here.
UPDATE (Apr 2019): In order to have a closer relationship between the GCP and G Suite worlds of Google Cloud, all G Suite Python code samples have been updated, replacing some of the older G Suite API client libraries with their equivalents from GCP. NOTE: using the newer libraries requires more initial code/effort from the developer thus will seem "less Pythonic." However, we will leave the code sample here with the original client libraries (deprecated but not shutdown yet) to be consistent with the video.
UPDATE (Aug 2016): The code has been modernized to use
oauth2client.tools.run_flow() instead of the deprecated oauth2client.tools.run_flow(). You can read more about that change here.UPDATE (Jun 2016): Updated to Python 2.7 & 3.3+ and Drive API v3.
Introduction
In this final installment of a (currently) two-part series introducing Python developers to building on Google APIs, we'll extend from the simple API example from the first post (part 1) just over a month ago. Those first snippets showed some skeleton code and a short real working sample that demonstrate accessing a public (Google) API with an API key (that queried public Google+ posts). An API key however, does not grant applications access to authorized data.Authorized data, including user information such as personal files on Google Drive and YouTube playlists, require additional security steps before access is granted. Sharing of and hardcoding credentials such as usernames and passwords is not only insecure, it's also a thing of the past. A more modern approach leverages token exchange, authenticated API calls, and standards such as OAuth2.
In this post, we'll demonstrate how to use Python to access authorized Google APIs using OAuth2, specifically listing the files (and folders) in your Google Drive. In order to better understand the example, we strongly recommend you check out the OAuth2 guides (general OAuth2 info, OAuth2 as it relates to Python and its client library) in the documentation to get started.
The docs describe the OAuth2 flow: making a request for authorized access, having the user grant access to your app, and obtaining a(n access) token with which to sign and make authorized API calls with. The steps you need to take to get started begin nearly the same way as for simple API access. The process diverges when you arrive on the Credentials page when following the steps below.
Google API access
In order to Google API authorized access, follow these instructions (the first three of which are roughly the same for simple API access):- Go to the Google Developers Console and login.
- Use your Gmail or Google credentials; create an account if needed
- Click "Create a Project" from pulldown under your username (at top)
- Enter a Project Name (mutable, human-friendly string only used in the console)
- Enter a Project ID (immutable, must be unique and not already taken)
- Once project has been created, enable APIs you wish to use
- You can toggle on any API(s) that support(s) simple or authorized API access.
- For the code example below, we use the Google Drive API.
- Other ideas: YouTube Data API, Google Sheets API, etc.
- Find more APIs (and version#s which you need) at the OAuth Playground.
- Select "Credentials" in left-nav
- Click "Create credentials" and select OAuth client ID
- In the new dialog, select your application type — we're building a command-line script which is an "Installed application"
- In the bottom part of that same dialog, specify the type of installed application; choose "Other" (cmd-line scripts are not web nor mobile)
- Click "Create Client ID" to generate your credentials
- Finally, click "Download JSON" to save the new credentials to your computer... perhaps choose a shorter name like "client_secret.json" or "client_id.json"
Accessing Google APIs from Python
In order to access authorized Google APIs from Python, you still need the Google APIs Client Library for Python, so in this case, do follow those installation instructions from part 1.We will again use the apiclient.discovery.build() function, which is what we need to create a service endpoint for interacting with an API, authorized or otherwise. However, for authorized data access, we need additional resources, namely the httplib2 and oauth2client packages. Here are the first five lines of the new boilerplate code for authorized access:
from __future__ import print_function
from googleapiclient import discoverySCOPES is a critical variable: it represents the set of scopes of authorization an app wants to obtain (then access) on behalf of user(s). What's does a scope look like?
from httplib2 import Http
from oauth2client import file, client, tools
SCOPES = # one or more scopes (strings)
Each scope is a single character string, specifically a URL. Here are some examples:
- 'https://www.googleapis.com/auth/plus.me'— access your personal Google+ settings
- 'https://www.googleapis.com/auth/drive.metadata.readonly'— read-only access your Google Drive file or folder metadata
- 'https://www.googleapis.com/auth/youtube'— access your YouTube playlists and other personal information
SCOPES = 'https://www.googleapis.com/auth/plus.me https://www.googleapis.com/auth/youtube'That is space-delimited and made tiny by me so it doesn't wrap in a regular-sized browser window; or it could be an easier-to-read, non-tiny, and non-wrapped tuple:
SCOPES = (
'https://www.googleapis.com/auth/plus.me',
'https://www.googleapis.com/auth/youtube',
)
Our example command-line script will just list the files on your Google Drive, so we only need the read-only Drive metadata scope, meaning our SCOPES variable will be just this:
SCOPES = 'https://www.googleapis.com/auth/drive.metadata.readonly'The next section of boilerplate represents the security code:
store = file.Storage('storage.json')
creds = store.get()
if not creds or creds.invalid:
flow = client.flow_from_clientsecrets('client_secret.json', SCOPES)
creds = tools.run_flow(flow, store)
Once the user has authorized access to their personal data by your app, a special "access token" is given to your app. This precious resource must be stored somewhere local for the app to use. In our case, we'll store it in a file called "storage.json". The lines setting the store and creds variables are attempting to get a valid access token with which to make an authorized API call.If the credentials are missing or invalid, such as being expired, the authorization flow (using the client secret you downloaded along with a set of requested scopes) must be created (by client.flow_from_clientsecrets()) and executed (by tools.run_flow()) to ensure possession of valid credentials. The client_secret.json file is the credentials file you saved when you clicked "Download JSON" from the DevConsole after you've created your OAuth2 client ID.
If you don't have credentials at all, the user much explicitly grant permission — I'm sure you've all seen the OAuth2 dialog describing the type of access an app is requesting (remember those scopes?). Once the user clicks "Accept" to grant permission, a valid access token is returned and saved into the storage file (because you passed a handle to it when you called tools.run_flow()).
Note: tools.run() deprecated by tools.run_flow()You may have seen usage of the older tools.run() function, but it has been deprecated by tools.run_flow(). We explain this in more detail in another blogpost specifically geared towards migration. |
Once the user grants access and valid credentials are saved, you can create one or more endpoints to the secure service(s) desired with apiclient.discovery.build(), just like with simple API access. Its call will look slightly different, mainly that you need to sign your HTTP requests with your credentials rather than passing an API key:
In our example, we're going to list your files and folders in your Google Drive, so for API, use the string 'drive'. The API is currently on version 3 so use 'v3' for VERSION:
DRIVE = discovery.build('drive', 'v3', http=creds.authorize(Http()))
Going back to our working example, once you have an established service endpoint, you can use the list() method of the files service to request the file data:
files = DRIVE.files().list().execute().get('files', [])
If there's any data to read, the response dict will contain an iterable of files that we can loop over (or default to an empty list so the loop doesn't fail), displaying file names and types:
for f in files:
print(f['name'], f['mimeType'])
Conclusion
To find out more about the input parameters as well as all the fields that are in the response, take a look at the docs for files().list(). For more information on what other operations you can execute with the Google Drive API, take a look at the reference docs and check out the companion video for this code sample. That's it!Below is the entire script for your convenience:
'''When you run it, you should see pretty much what you'd expect, a list of file or folder names followed by their MIMEtypes — I named my script drive_list.py:
drive_list.py -- Google Drive API authorized demo
updated Aug 2016 by +WesleyChun/@wescpy
'''
from __future__ import print_function
from apiclient import discovery
from httplib2 import Http
from oauth2client import file, client, tools
SCOPES = 'https://www.googleapis.com/auth/drive.readonly.metadata'
store = file.Storage('storage.json')
creds = store.get()
if not creds or creds.invalid:
flow = client.flow_from_clientsecrets('client_secret.json', SCOPES)
creds = tools.run_flow(flow, store)
DRIVE = discovery.build('drive', 'v3', http=creds.authorize(Http()))
files = DRIVE.files().list().execute().get('files', [])
for f in files:
print(f['name'], f['mimeType'])
$ python3 drive_list.py
Google Maps demo application/vnd.google-apps.spreadsheet
Overview of Google APIs - Sep 2014 application/vnd.google-apps.presentation
tiresResearch.xls application/vnd.google-apps.spreadsheet
6451_Core_Python_Schedule.doc application/vnd.google-apps.document
out1.txt application/vnd.google-apps.document
tiresResearch.xls application/vnd.ms-excel
6451_Core_Python_Schedule.doc application/msword
out1.txt text/plain
Maps and Sheets demo application/vnd.google-apps.spreadsheet
ProtoRPC Getting Started Guide application/vnd.google-apps.document
gtaskqueue-1.0.2_public.tar.gz application/x-gzip
Pull Queues application/vnd.google-apps.folder
gtaskqueue-1.0.1_public.tar.gz application/x-gzip
appengine-java-sdk.zip application/zip
taskqueue.py text/x-python-script
Google Apps Security Whitepaper 06/10/2010.pdf application/pdf
Obviously your output will be different, depending on what files are in your Google Drive. But that's it... hope this is useful. You can now customize this code for your own needs and/or to access other Google APIs. Thanks for reading!EXTRA CREDIT: To test your skills, add functionality to this code that also displays the last modified timestamp, the file (byte)size, and perhaps shave the MIMEtype a bit as it's slightly harder to read in its entirety... perhaps take just the final path element? One last challenge: in the output above, we have both Microsoft Office documents as well as their auto-converted versions for Google Apps... perhaps only show the filename once and have a double-entry for the filetypes!
Andre Roberge: Friendly Mu
Kushal Das: Indian police attacked university campuses on government order

Yesterday, Indian police attacked protesting students across different university campuses. They fired tear gas shells inside of libraries; they lit buses on fire and then told that the students did it. They broke into a Mosque and beat up students there.
The Internet has been shut down in Kashmir for over 130 days, and now few more states + different smaller parts of the country are having the same.
Search for #JamiaProtest or #SOSJamia on twitter to see what is going on in India. I asked to my around 5k followers, to reply if they can see our tweets (only around 5 replied via the original tweet).
Trigger warning (The following tweets shows police brutality)
I have curated a few tweets for you, please see these (if possible) and then share those.
- Police + goons (posing as police) beating up the girls https://twitter.com/amirul_alig/status/1206273340194746368
- Police attacked the library where students were studying https://twitter.com/i_kathayat/status/1206271022976008194
- Police attacking Jamia Milia University library https://twitter.com/imMAK02/status/1206213159981203458
- Police vandalizing university https://twitter.com/amirul_alig/status/1206273340194746368
- Police firing tear gases at AMU https://twitter.com/imMAK02/status/1206244102641049600
- Police firing their guns https://twitter.com/imMAK02/status/1206233908083187712
- Police vandalizing public property https://twitter.com/imMAK02/status/1206259162969038848
- Police breaking the gate of Aligarh Muslim University https://twitter.com/imMAK02/status/1206261850943311873
- Police putting fire on a bus https://twitter.com/DesiPoliticks/status/1206222796927438848
- Police attacking AMU https://twitter.com/tariq_shameem/status/1206259086796279810
Why am I writing this in my blog (maybe you are reading it on a technical planet)?
Most people are ignorant about the fascist regime in India, and the IT industry (including us) mostly tries to pretend that everything is perfect. I hope at least a few more around will read the tweets linked from this post and also watch the videos. I hope you will share those in your social circles. To stop fascists, we have to rise together.

Btw, you should at least read this story from New Yorker on how the fascist government is attacking the fellow citizens.
To know about the reason behind the current protest, read this story showing the similarities between Nazi Germany and current Indian government.
Top most drawing credit: I am yet to find the original artist, I will update when I find the name.
Mike Driscoll: PyDev of the Week: Ted Petrou
This week we welcome Ted Petrou (@TedPetrou) as our PyDev of the Week! Ted is the author of the Pandas Cookbook and also teaches Pandas in several courses on Udemy. Let’s take some time to get to know Ted better!
Can you tell us a little about yourself (hobbies, education, etc):
I graduated with a masters degree in statistics from Rice University in Houston, Texas in 2006. During my degree, I never heard the phrase “machine learning” uttered even once and it was several years before the field of data science became popular. I had entered the program pursuing a Ph.D with just six other students. Although statistics was a highly viable career at the time, it wasn’t nearly as popular as it is today.
After limping out of the program with a masters degree, I looked into the fields of actuarial science, became a professional poker play, taught high school math, built reports with SQL and Excel VBA as a financial analyst before becoming a data scientist at Schlumberger. During my stint as a data scientist, I started the meetup group Houston Data Science where I gave tutorials on various Python data science topics. Once I accumulated enough material, I started my company Dunder Data, teaching data science full time.
Why did you start using Python?
I began using Python when I took an introductory course offered by Rice University on coursera.org in 2013 when I was teaching high school math. I had done quite a bit of programming prior to that, but had never heard of Python before. It was a great course where we built a new game each week.
What other programming languages do you know and which is your favorite?
I began programming on a TI-82 calculator about 22 years ago. There was a minimal built-in language that my friends and I would use to build games. I remember making choose-your-own adventure games using the menu command. I took classes in C and Java in college and worked with R as a graduate student. A while later, I learned enough HTML and JavaScript to build basic websites. I also know SQL quite well and have done some work in Excel VBA.
My favorite language is Python, but I have no emotional attachment to it. I’d actually prefer to use a language that is statically typed, but I don’t have much of a choice as the demand for Python is increasing.
What projects are you working on now?
Outside of building material for my courses, I have two major projects, Dexplo and Dexplot, that are in current development. Dexplo is a data analysis library similar to pandas with the goal of having simpler syntax, better performance, one obvious way to perform common tasks, and more functionality. Dexplot is similar to seaborn with similar goals.
Which Python libraries are your favorite (core or 3rd party)?
I really enjoy using scikit-learn. The uniformity of the estimator object makes it easy to use. The recent introduction of the ColumnTransformer and the upgrade to the OneHotEncoder have really improved the library.
I see you are an author. How did that come about?
I contacted both O’Reilly and Packt Publishing with book ideas. O’Reilly offered me an online course and Packt needed an author for Pandas Cookbook. I really wanted to write a book and already had lots of material available from teaching my data science bootcamp so I went with Packt.
Do you have any advice for other aspiring authors?
It really helps to teach the material before you publish it. The feedback you get from the students is invaluable to making improvements. You can see firsthand what works and what needs to be changed.
What’s the origin story for your company, Dunder Data?
During my time as a data scientist at Schlumberger, I participated in several weeks of poorly taught corporate training. This experience motivated me to start creating tutorials. I started the Houston Data Science meetup group which helped lay the foundation for my Dunder Data. Many people ask if the “Dunder” is related to Dunder Mifflin, the paper company from the popular TV show, The Office. The connection is coincidental, as dunder refers to “magic” or “special” methods. The idea is that Dunder Data translates to “Magical Data”.
Thanks for doing the interview, Ted!
The post PyDev of the Week: Ted Petrou appeared first on The Mouse Vs. The Python.
Janusworx: #100DaysOfCode, Days 024, 025 & 026 – Watched Videos
Life has suddenly turned a little topsy turvy at work.
No time to work at stuff.
Keeping up my #100DaysOfCode streak by watching the course videos.
Learnt about the Itertools module, learnt decorators and error handling.
P.S.
Adding these posts back to the planet, because more than a couple of you, kind folk, have been missing me.
Real Python: Python Statistics Fundamentals: How to Describe Your Data
In the era of big data and artificial intelligence, data science and machine learning have become essential in many fields of science and technology. A necessary aspect of working with data is the ability to describe, summarize, and represent data visually. Python statistics libraries are comprehensive, popular, and widely used tools that will assist you in working with data.
In this tutorial, you’ll learn:
- What numerical quantities you can use to describe and summarize your datasets
- How to calculate descriptive statistics in pure Python
- How to get descriptive statistics with available Python libraries
- How to visualize your datasets
Free Bonus:Click here to download 5 Python + Matplotlib examples with full source code that you can use as a basis for making your own plots and graphics.
Understanding Descriptive Statistics
Descriptive statistics is about describing and summarizing data. It uses two main approaches:
- The quantitative approach describes and summarizes data numerically.
- The visual approach illustrates data with charts, plots, histograms, and other graphs.
You can apply descriptive statistics to one or many datasets or variables. When you describe and summarize a single variable, you’re performing univariate analysis. When you search for statistical relationships among a pair of variables, you’re doing a bivariate analysis. Similarly, a multivariate analysis is concerned with multiple variables at once.
Types of Measures
In this tutorial, you’ll learn about the following types of measures in descriptive statistics:
- Central tendency tells you about the centers of the data. Useful measures include the mean, median, and mode.
- Variability tells you about the spread of the data. Useful measures include variance and standard deviation.
- Correlation or joint variability tells you about the relation between a pair of variables in a dataset. Useful measures include covariance and the correlation coefficient.
You’ll learn how to understand and calculate these measures with Python.
Population and Samples
In statistics, the population is a set of all elements or items that you’re interested in. Populations are often vast, which makes them inappropriate for collecting and analyzing data. That’s why statisticians usually try to make some conclusions about a population by choosing and examining a representative subset of that population.
This subset of a population is called a sample. Ideally, the sample should preserve the essential statistical features of the population to a satisfactory extent. That way, you’ll be able to use the sample to glean conclusions about the population.
Outliers
An outlier is a data point that differs significantly from the majority of the data taken from a sample or population. There are many possible causes of outliers, but here are a few to start you off:
- Natural variation in data
- Change in the behavior of the observed system
- Errors in data collection
Data collection errors are a particularly prominent cause of outliers. For example, the limitations of measurement instruments or procedures can mean that the correct data is simply not obtainable. Other errors can be caused by miscalculations, data contamination, human error, and more.
There isn’t a precise mathematical definition of outliers. You have to rely on experience, knowledge about the subject of interest, and common sense to determine if a data point is an outlier and how to handle it.
Choosing Python Statistics Libraries
There are many Python statistics libraries out there for you to work with, but in this tutorial, you’ll be learning about some of the most popular and widely used ones:
Python’s
statisticsis a built-in Python library for descriptive statistics. You can use it if your datasets are not too large or if you can’t rely on importing other libraries.NumPy is a third-party library for numerical computing, optimized for working with single- and multi-dimensional arrays. Its primary type is the array type called
ndarray. This library contains many routines for statistical analysis.SciPy is a third-party library for scientific computing based on NumPy. It offers additional functionality compared to NumPy, including
scipy.statsfor statistical analysis.Pandas is a third-party library for numerical computing based on NumPy. It excels in handling labeled one-dimensional (1D) data with
Seriesobjects and two-dimensional (2D) data withDataFrameobjects.Matplotlib is a third-party library for data visualization. It works well in combination with NumPy, SciPy, and Pandas.
Note that, in many cases, Series and DataFrame objects can be used in place of NumPy arrays. Often, you might just pass them to a NumPy or SciPy statistical function. In addition, you can get the unlabeled data from a Series or DataFrame as a np.ndarray object by calling .values or .to_numpy().
Getting Started With Python Statistics Libraries
The built-in Python statistics library has a relatively small number of the most important statistics functions. The official documentation is a valuable resource to find the details. If you’re limited to pure Python, then the Python statistics library might be the right choice.
A good place to start learning about NumPy is the official User Guide, especially the quickstart and basics sections. The official reference can help you refresh your memory on specific NumPy concepts. While you read this tutorial, you might want to check out the statistics section and the official scipy.stats reference as well.
Note:
To learn more about NumPy, check out these resources:
If you want to learn Pandas, then the official Getting Started page is an excellent place to begin. The introduction to data structures can help you learn about the fundamental data types, Series and DataFrame. Likewise, the excellent official introductory tutorial aims to give you enough information to start effectively using Pandas in practice.
Note:
To learn more about Pandas, check out these resources:
matplotlib has a comprehensive official User’s Guide that you can use to dive into the details of using the library. Anatomy of Matplotlib is an excellent resource for beginners who want to start working with matplotlib and its related libraries.
Note:
To learn more about data visualization, check out these resources:
Let’s start using these Python statistics libraries!
Calculating Descriptive Statistics
Start by importing all the packages you’ll need:
>>> importmath>>> importstatistics>>> importnumpyasnp>>> importscipy.stats>>> importpandasaspdThese are all the packages you’ll need for Python statistics calculations. Usually, you won’t use Python’s built-in math package, but it’ll be useful in this tutorial. Later, you’ll import matplotlib.pyplot for data visualization.
Let’s create some data to work with. You’ll start with Python lists that contain some arbitrary numeric data:
>>> x=[8.0,1,2.5,4,28.0]>>> x_with_nan=[8.0,1,2.5,math.nan,4,28.0]>>> x[8.0, 1, 2.5, 4, 28.0]>>> x_with_nan[8.0, 1, 2.5, nan, 4, 28.0]Now you have the lists x and x_with_nan. They’re almost the same, with the difference that x_with_nan contains a nan value. It’s important to understand the behavior of the Python statistics routines when they come across a not-a-number value (nan). In data science, missing values are common, and you’ll often replace them with nan.
Note: How do you get a nan value?
In Python, you can use any of the following:
You can use all of these functions interchangeably:
>>> math.isnan(np.nan),np.isnan(math.nan)(True, True)>>> math.isnan(y_with_nan[3]),np.isnan(y_with_nan[3])(True, True)You can see that the functions are all equivalent. However, please keep in mind that comparing two nan values for equality returns False. In other words, math.nan == math.nan is False!
Now, create np.ndarray and pd.Series objects that correspond to x and x_with_nan:
>>> y,y_with_nan=np.array(x),np.array(x_with_nan)>>> z,z_with_nan=pd.Series(x),pd.Series(x_with_nan)>>> yarray([ 8. , 1. , 2.5, 4. , 28. ])>>> y_with_nanarray([ 8. , 1. , 2.5, nan, 4. , 28. ])>>> z0 8.01 1.02 2.53 4.04 28.0dtype: float64>>> z_with_nan0 8.01 1.02 2.53 NaN4 4.05 28.0dtype: float64You now have two NumPy arrays (y and y_with_nan) and two Pandas Series (z and z_with_nan). All of these are 1D sequences of values.
Note: Although you’ll use lists throughout this tutorial, please keep in mind that, in most cases, you can use tuples in the same way.
You can optionally specify a label for each value in z and z_with_nan.
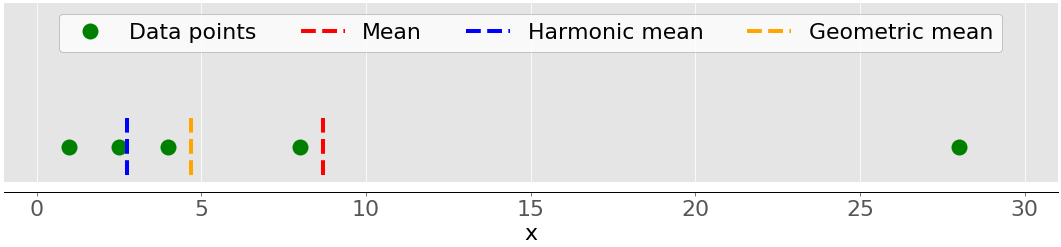
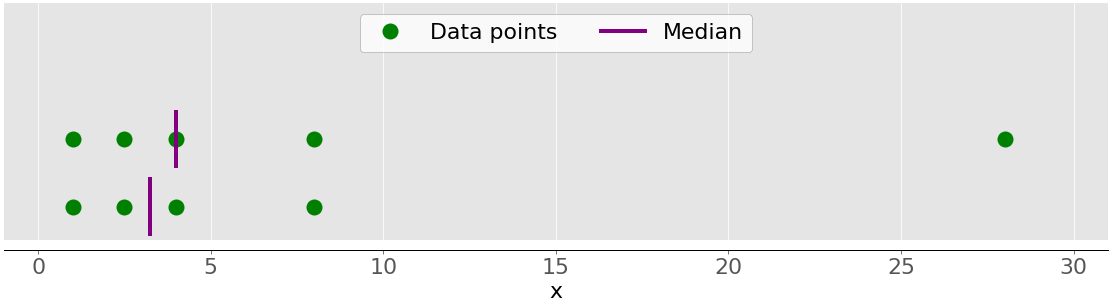
Measures of Central Tendency
The measures of central tendency show the central or middle values of datasets. There are several definitions of what’s considered to be the center of a dataset. In this tutorial, you’ll learn how to identify and calculate these measures of central tendency:
- Mean
- Weighted mean
- Geometric mean
- Harmonic mean
- Median
- Mode
Mean
The sample mean, also called the sample arithmetic mean or simply the average, is the arithmetic average of all the items in a dataset. The mean of a dataset 𝑥 is mathematically expressed as Σᵢ𝑥ᵢ/𝑛, where 𝑖 = 1, 2, …, 𝑛. In other words, it’s the sum of all the elements 𝑥ᵢ divided by the number of items in the dataset 𝑥.
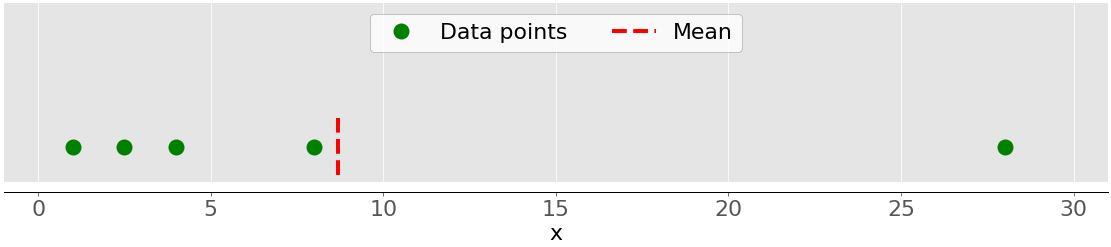
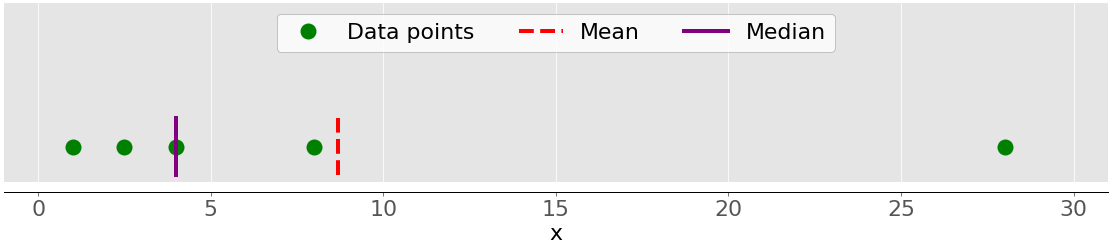
This figure illustrates the mean of a sample with five data points:
The green dots represent the data points 1, 2.5, 4, 8, and 28. The red dashed line is their mean, or (1 + 2.5 + 4 + 8 + 28) / 5 = 8.7.
You can calculate the mean with pure Python using sum() and len(), without importing libraries:
>>> mean_=sum(x)/len(x)>>> mean_8.7Although this is clean and elegant, you can also apply built-in Python statistics functions:
>>> mean_=statistics.mean(x)>>> mean_8.7>>> mean_=statistics.fmean(x)>>> mean_8.7You’ve called the functions mean() and fmean() from the built-in Python statistics library and got the same result as you did with pure Python. fmean() is introduced in Python 3.8 as a faster alternative to mean(). It always returns a floating-point number.
However, if there are nan values among your data, then statistics.mean() and statistics.fmean() will return nan as the output:
>>> mean_=statistics.mean(x_with_nan)>>> mean_nan>>> mean_=statistics.fmean(x_with_nan)>>> mean_nanThis result is consistent with the behavior of sum(), because sum(x_with_nan) also returns nan.
If you use NumPy, then you can get the mean with np.mean():
>>> mean_=np.mean(y)>>> mean_8.7In the example above, mean() is a function, but you can use the corresponding method .mean() as well:
>>> mean_=y.mean()>>> mean_8.7The function mean() and method .mean() from NumPy return the same result as statistics.mean(). This is also the case when there are nan values among your data:
>>> np.mean(y_with_nan)nan>>> y_with_nan.mean()nanYou often don’t need to get a nan value as a result. If you prefer to ignore nan values, then you can use np.nanmean():
>>> np.nanmean(y_with_nan)8.7nanmean() simply ignores all nan values. It returns the same value as mean() if you were to apply it to the dataset without the nan values.
pd.Series objects also have the method .mean():
>>> mean_=z.mean()>>> mean_8.7As you can see, it’s used similarly as in the case of NumPy. However, .mean() from Pandas ignores nan values by default:
>>> z_with_nan.mean()8.7This behavior is the result of the default value of the optional parameter skipna. You can change this parameter to modify the behavior.
Weighted Mean
The weighted mean, also called the weighted arithmetic mean or weighted average, is a generalization of the arithmetic mean that enables you to define the relative contribution of each data point to the result.
You define one weight 𝑤ᵢ for each data point 𝑥ᵢ of the dataset 𝑥, where 𝑖 = 1, 2, …, 𝑛 and 𝑛 is the number of items in 𝑥. Then, you multiply each data point with the corresponding weight, sum all the products, and divide the obtained sum with the sum of weights: Σᵢ(𝑤ᵢ𝑥ᵢ) / Σᵢ𝑤ᵢ.
Note: It’s convenient (and usually the case) that all weights are nonnegative, 𝑤ᵢ ≥ 0, and that their sum is equal to one, or Σᵢ𝑤ᵢ = 1.
The weighted mean is very handy when you need the mean of a dataset containing items that occur with given relative frequencies. For example, say that you have a set in which 20% of all items are equal to 2, 50% of the items are equal to 4, and the remaining 30% of the items are equal to 8. You can calculate the mean of such a set like this:
>>> 0.2*2+0.5*4+0.3*84.8Here, you take the frequencies into account with the weights. With this method, you don’t need to know the total number of items.
You can implement the weighted mean in pure Python by combining sum() with either range() or zip():
>>> x=[8.0,1,2.5,4,28.0]>>> w=[0.1,0.2,0.3,0.25,0.15]>>> wmean=sum(w[i]*x[i]foriinrange(len(x)))/sum(w)>>> wmean6.95>>> wmean=sum(x_*w_for(x_,w_)inzip(x,w))/sum(w)>>> wmean6.95Again, this is a clean and elegant implementation where you don’t need to import any libraries.
However, if you have large datasets, then NumPy is likely to provide a better solution. You can use np.average() to get the weighted mean of NumPy arrays or Pandas Series:
>>> y,z,w=np.array(x),pd.Series(x),np.array(w)>>> wmean=np.average(y,weights=w)>>> wmean6.95>>> wmean=np.average(z,weights=w)>>> wmean6.95The result is the same as in the case of the pure Python implementation. You can also use this method on ordinary lists and tuples.
Another solution is to use the element-wise product w * y with np.sum() or .sum():
>>> (w*y).sum()/w.sum()6.95That’s it! You’ve calculated the weighted mean.
However, be careful if your dataset contains nan values:
>>> w=np.array([0.1,0.2,0.3,0.0,0.2,0.1])>>> (w*y_with_nan).sum()/w.sum()nan>>> np.average(y_with_nan,weights=w)nan>>> np.average(z_with_nan,weights=w)nanIn this case, average() returns nan, which is consistent with np.mean().
Harmonic Mean
The harmonic mean is the reciprocal of the mean of the reciprocals of all items in the dataset: 𝑛 / Σᵢ(1/𝑥ᵢ), where 𝑖 = 1, 2, …, 𝑛 and 𝑛 is the number of items in the dataset 𝑥. One variant of the pure Python implementation of the harmonic mean is this:
>>> hmean=len(x)/sum(1/itemforiteminx)>>> hmean2.7613412228796843It’s quite different from the value of the arithmetic mean for the same data x, which you calculated to be 8.7.
You can also calculate this measure with statistics.harmonic_mean():
>>> hmean=statistics.harmonic_mean(x)>>> hmean2.7613412228796843The example above shows one implementation of statistics.harmonic_mean(). If you have a nan value in a dataset, then it’ll return nan. If there’s at least one 0, then it’ll return 0. If you provide at least one negative number, then you’ll get statistics.StatisticsError:
>>> statistics.harmonic_mean(x_with_nan)nan>>> statistics.harmonic_mean([1,0,2])0>>> statistics.harmonic_mean([1,2,-2])# Raises StatisticsErrorKeep these three scenarios in mind when you’re using this method!
A third way to calculate the harmonic mean is to use scipy.stats.hmean():
>>> scipy.stats.hmean(y)2.7613412228796843>>> scipy.stats.hmean(z)2.7613412228796843Again, this is a pretty straightforward implementation. However, if your dataset contains nan, 0, a negative number, or anything but positive numbers, then you’ll get a ValueError!
Geometric Mean
The geometric mean is the 𝑛-th root of the product of all 𝑛 elements 𝑥ᵢ in a dataset 𝑥: ⁿ√(Πᵢ𝑥ᵢ), where 𝑖 = 1, 2, …, 𝑛. The following figure illustrates the arithmetic, harmonic, and geometric means of a dataset:
Again, the green dots represent the data points 1, 2.5, 4, 8, and 28. The red dashed line is the mean. The blue dashed line is the harmonic mean, and the yellow dashed line is the geometric mean.
You can implement the geometric mean in pure Python like this:
>>> gmean=1>>> foriteminx:... gmean*=item...>>> gmean**=1/len(x)>>> gmean4.677885674856041As you can see, the value of the geometric mean, in this case, differs significantly from the values of the arithmetic (8.7) and harmonic (2.76) means for the same dataset x.
Python 3.8 introduced statistics.geometric_mean(), which converts all values to floating-point numbers and returns their geometric mean:
>>> gmean=statistics.geometric_mean(x)>>> gmean4.67788567485604You’ve got the same result as in the previous example, but with a minimal rounding error.
If you pass data with nan values, then statistics.geometric_mean() will behave like most similar functions and return nan:
>>> gmean=statistics.geometric_mean(x_with_nan)>>> gmeannanIndeed, this is consistent with the behavior of statistics.mean(), statistics.fmean(), and statistics.harmonic_mean(). If there’s a zero or negative number among your data, then statistics.geometric_mean() will raise the statistics.StatisticsError.
You can also get the geometric mean with scipy.stats.gmean():
>>> scipy.stats.gmean(y)4.67788567485604>>> scipy.stats.gmean(z)4.67788567485604You obtained the same result as with the pure Python implementation.
If you have nan values in a dataset, then gmean() will return nan. If there’s at least one 0, then it’ll return 0.0 and give a warning. If you provide at least one negative number, then you’ll get nan and the warning.
Median
The sample median is the middle element of a sorted dataset. The dataset can be sorted in increasing or decreasing order. If the number of elements 𝑛 of the dataset is odd, then the median is the value at the middle position: 0.5(𝑛 + 1). If 𝑛 is even, then the median is the arithmetic mean of the two values in the middle, that is, the items at the positions 0.5𝑛 and 0.5𝑛 + 1.
For example, if you have the data points 2, 4, 1, 8, and 9, then the median value is 4, which is in the middle of the sorted dataset (1, 2, 4, 8, 9). If the data points are 2, 4, 1, and 8, then the median is 3, which is the average of the two middle elements of the sorted sequence (2 and 4). The following figure illustrates this:
The data points are the green dots, and the purple lines show the median for each dataset. The median value for the upper dataset (1, 2.5, 4, 8, and 28) is 4. If you remove the outlier 28 from the lower dataset, then the median becomes the arithmetic average between 2.5 and 4, which is 3.25.
The figure below shows both the mean and median of the data points 1, 2.5, 4, 8, and 28:
Again, the mean is the red dashed line, while the median is the purple line.
The main difference between the behavior of the mean and median is related to dataset outliers or extremes. The mean is heavily affected by outliers, but the median only depends on outliers either slightly or not at all. Consider the following figure:
The upper dataset again has the items 1, 2.5, 4, 8, and 28. Its mean is 8.7, and the median is 5, as you saw earlier. The lower dataset shows what’s going on when you move the rightmost point with the value 28:
- If you increase its value (move it to the right), then the mean will rise, but the median value won’t ever change.
- If you decrease its value (move it to the left), then the mean will drop, but the median will remain the same until the value of the moving point is greater than or equal to 4.
You can compare the mean and median as one way to detect outliers and asymmetry in your data. Whether the mean value or the median value is more useful to you depends on the context of your particular problem.
Here is one of many possible pure Python implementations of the median:
>>> n=len(x)>>> ifn%2:... median_=sorted(x)[round(0.5*(n-1))]... else:... x_ord,index=sorted(x),round(0.5*n)... median_=0.5*(x_ord[index-1]+x_ord[index])...>>> median_4Two most important steps of this implementation are as follows:
- Sorting the elements of the dataset
- Finding the middle element(s) in the sorted dataset
You can get the median with statistics.median():
>>> median_=statistics.median(x)>>> median_4>>> median_=statistics.median(x[:-1])>>> median_3.25The sorted version of x is [1, 2.5, 4, 8.0, 28.0], so the element in the middle is 4. The sorted version of x[:-1], which is x without the last item 28.0, is [1, 2.5, 4, 8.0]. Now, there are two middle elements, 2.5 and 4. Their average is 3.25.
median_low() and median_high() are two more functions related to the median in the Python statistics library. They always return an element from the dataset:
- If the number of elements is odd, then there’s a single middle value, so these functions behave just like
median(). - If the number of elements is even, then there are two middle values. In this case,
median_low()returns the lower andmedian_high()the higher middle value.
You can use these functions just as you’d use median():
>>> statistics.median_low(x[:-1])2.5>>> statistics.median_high(x[:-1])4Again, the sorted version of x[:-1] is [1, 2.5, 4, 8.0]. The two elements in the middle are 2.5 (low) and 4 (high).
Unlike most other functions from the Python statistics library, median(), median_low(), and median_high() don’t return nan when there are nan values among the data points:
>>> statistics.median(x_with_nan)6.0>>> statistics.median_low(x_with_nan)4>>> statistics.median_high(x_with_nan)8.0Beware of this behavior because it might not be what you want!
You can also get the median with np.median():
>>> median_=np.median(y)>>> median_4.0>>> median_=np.median(y[:-1])>>> median_3.25You’ve obtained the same values with statistics.median() and np.median().
However, if there’s a nan value in your dataset, then np.median() issues the RuntimeWarning and returns nan. If this behavior is not what you want, then you can use nanmedian() to ignore all nan values:
>>> np.nanmedian(y_with_nan)4.0>>> np.nanmedian(y_with_nan[:-1])3.25The obtained results are the same as with statistics.median() and np.median() applied to the datasets x and y.
Pandas Series objects have the method .median() that ignores nan values by default:
>>> z.median()4.0>>> z_with_nan.median()4.0The behavior of .median() is consistent with .mean() in Pandas. You can change this behavior with the optional parameter skipna.
Mode
The sample mode is the value in the dataset that occurs most frequently. If there isn’t a single such value, then the set is multimodal since it has multiple modal values. For example, in the set that contains the points 2, 3, 2, 8, and 12, the number 2 is the mode because it occurs twice, unlike the other items that occur only once.
This is how you can get the mode with pure Python:
>>> u=[2,3,2,8,12]>>> mode_=max((u.count(item),item)foriteminset(u))[1]>>> mode_2You use u.count() to get the number of occurrences of each item in u. The item with the maximal number of occurrences is the mode. Note that you don’t have to use set(u). Instead, you might replace it with just u and iterate over the entire list.
Note:set(u) returns a Python set with all unique items in u. You can use this trick to optimize working with larger data, especially when you expect to see a lot of duplicates.
You can obtain the mode with statistics.mode() and statistics.multimode():
>>> mode_=statistics.mode(u)>>> mode_>>> mode_=statistics.multimode(u)>>> mode_[2]As you can see, mode() returned a single value, while multimode() returned the list that contains the result. This isn’t the only difference between the two functions, though. If there’s more than one modal value, then mode() raises StatisticsError, while multimode() returns the list with all modes:
>>> v=[12,15,12,15,21,15,12]>>> statistics.mode(v)# Raises StatisticsError>>> statistics.multimode(v)[12, 15]You should pay special attention to this scenario and be careful when you’re choosing between these two functions.
statistics.mode() and statistics.multimode() handle nan values as regular values and can return nan as the modal value:
>>> statistics.mode([2,math.nan,2])2>>> statistics.multimode([2,math.nan,2])[2]>>> statistics.mode([2,math.nan,0,math.nan,5])nan>>> statistics.multimode([2,math.nan,0,math.nan,5])[nan]In the first example above, the number 2 occurs twice and is the modal value. In the second example, nan is the modal value since it occurs twice, while the other values occur only once.
Note:statistics.multimode() is introduced in Python 3.8.
You can also get the mode with scipy.stats.mode():
>>> u,v=np.array(u),np.array(v)>>> mode_=scipy.stats.mode(u)>>> mode_ModeResult(mode=array([2]), count=array([2]))>>> mode_=scipy.stats.mode(v)>>> mode_ModeResult(mode=array([12]), count=array([3]))This function returns the object with the modal value and the number of times it occurs. If there are multiple modal values in the dataset, then only the smallest value is returned.
You can get the mode and its number of occurrences as NumPy arrays with dot notation:
>>> mode_.modearray([12])>>> mode_.countarray([3])This code uses .mode to return the smallest mode (12) in the array v and .count to return the number of times it occurs (3). scipy.stats.mode() is also flexible with nan values. It allows you to define desired behavior with the optional parameter nan_policy. This parameter can take on the values 'propagate', 'raise' (an error), or 'omit'.
Pandas Series objects have the method .mode() that handles multimodal values well and ignores nan values by default:
>>> u,v,w=pd.Series(u),pd.Series(v),pd.Series([2,2,math.nan])>>> u.mode()0 2dtype: int64>>> v.mode()0 121 15dtype: int64>>> w.mode()0 2.0dtype: float64As you can see, .mode() returns a new pd.Series that holds all modal values. If you want .mode() to take nan values into account, then just pass the optional argument dropna=False.
Measures of Variability
The measures of central tendency aren’t sufficient to describe data. You’ll also need the measures of variability that quantify the spread of data points. In this section, you’ll learn how to identify and calculate the following variability measures:
- Variance
- Standard deviation
- Skewness
- Percentiles
- Ranges
Variance
The sample variance quantifies the spread of the data. It shows numerically how far the data points are from the mean. You can express the sample variance of the dataset 𝑥 with 𝑛 elements mathematically as 𝑠² = Σᵢ(𝑥ᵢ − mean(𝑥))² / (𝑛 − 1), where 𝑖 = 1, 2, …, 𝑛 and mean(𝑥) is the sample mean of 𝑥. If you want to understand deeper why you divide the sum with 𝑛 − 1 instead of 𝑛, then you can dive deeper into Bessel’s correction.
The following figure shows you why it’s important to consider the variance when describing datasets:
There are two datasets in this figure:
- Green dots: This dataset has a smaller variance or a smaller average difference from the mean. It also has a smaller range or a smaller difference between the largest and smallest item.
- White dots: This dataset has a larger variance or a larger average difference from the mean. It also has a bigger range or a bigger difference between the largest and smallest item.
Note that these two datasets have the same mean and median, even though they appear to differ significantly. Neither the mean nor the median can describe this difference. That’s why you need the measures of variability.
Here’s how you can calculate the sample variance with pure Python:
>>> n=len(x)>>> mean_=sum(x)/n>>> var_=sum((item-mean_)**2foriteminx)/(n-1)>>> var_123.19999999999999This approach is sufficient and calculates the sample variance well. However, the shorter and more elegant solution is to call the existing function statistics.variance():
>>> var_=statistics.variance(x)>>> var_123.2You’ve obtained the same result for the variance as above. variance() can avoid calculating the mean if you provide the mean explicitly as the second argument: statistics.variance(x, mean_).
If you have nan values among your data, then statistics.variance() will return nan:
>>> statistics.variance(x_with_nan)nanThis behavior is consistent with mean() and most other functions from the Python statistics library.
You can also calculate the sample variance with NumPy. You should use the function np.var() or the corresponding method .var():
>>> var_=np.var(y,ddof=1)>>> var_123.19999999999999>>> var_=y.var(ddof=1)>>> var_123.19999999999999It’s very important to specify the parameter ddof=1. That’s how you set the delta degrees of freedom to 1. This parameter allows the proper calculation of 𝑠², with (𝑛 − 1) in the denominator instead of 𝑛.
If you have nan values in the dataset, then np.var() and .var() will return nan:
>>> np.var(y_with_nan,ddof=1)nan>>> y_with_nan.var(ddof=1)nanThis is consistent with np.mean() and np.average(). If you want to skip nan values, then you should use np.nanvar():
>>> np.nanvar(y_with_nan,ddof=1)123.19999999999999np.nanvar() ignores nan values. It also needs you to specify ddof=1.
pd.Series objects have the method .var() that skips nan values by default:
>>> z.var(ddof=1)123.19999999999999>>> z_with_nan.var(ddof=1)123.19999999999999It also has the parameter ddof, but its default value is 1, so you can omit it. If you want a different behavior related to nan values, then use the optional parameter skipna.
You calculate the population variance similarly to the sample variance. However, you have to use 𝑛 in the denominator instead of 𝑛 − 1: Σᵢ(𝑥ᵢ − mean(𝑥))² / 𝑛. In this case, 𝑛 is the number of items in the entire population. You can get the population variance similar to the sample variance, with the following differences:
- Replace
(n - 1)withnin the pure Python implementation. - Use
statistics.pvariance()instead ofstatistics.variance(). - Specify the parameter
ddof=0if you use NumPy or Pandas. In NumPy, you can omitddofbecause its default value is0.
Note that you should always be aware of whether you’re working with a sample or the entire population whenever you’re calculating the variance!
Standard Deviation
The sample standard deviation is another measure of data spread. It’s connected to the sample variance, as standard deviation, 𝑠, is the positive square root of the sample variance. The standard deviation is often more convenient than the variance because it has the same unit as the data points. Once you get the variance, you can calculate the standard deviation with pure Python:
>>> std_=var_**0.5>>> std_11.099549540409285Although this solution works, you can also use statistics.stdev():
>>> std_=statistics.stdev(x)>>> std_11.099549540409287Of course, the result is the same as before. Like variance(), stdev() doesn’t calculate the mean if you provide it explicitly as the second argument: statistics.stdev(x, mean_).
You can get the standard deviation with NumPy in almost the same way. You can use the function std() and the corresponding method .std() to calculate the standard deviation. If there are nan values in the dataset, then they’ll return nan. To ignore nan values, you should use np.nanstd(). You use std(), .std(), and nanstd() from NumPy as you would use var(), .var(), and nanvar():
>>> np.std(y,ddof=1)11.099549540409285>>> y.std(ddof=1)11.099549540409285>>> np.std(y_with_nan,ddof=1)nan>>> y_with_nan.std(ddof=1)nan>>> np.nanstd(y_with_nan,ddof=1)11.099549540409285Don’t forget to set the delta degrees of freedom to 1!
pd.Series objects also have the method .std() that skips nan by default:
>>> z.std(ddof=1)11.099549540409285>>> z_with_nan.std(ddof=1)11.099549540409285The parameter ddof defaults to 1, so you can omit it. Again, if you want to treat nan values differently, then apply the parameter skipna.
The population standard deviation refers to the entire population. It’s the positive square root of the population variance. You can calculate it just like the sample standard deviation, with the following differences:
- Find the square root of the population variance in the pure Python implementation.
- Use
statistics.pstdev()instead ofstatistics.stdev(). - Specify the parameter
ddof=0if you use NumPy or Pandas. In NumPy, you can omitddofbecause its default value is0.
As you can see, you can determine the standard deviation in Python, NumPy, and Pandas in almost the same way as you determine the variance. You use different but analogous functions and methods with the same arguments.
Skewness
The sample skewness measures the asymmetry of a data sample.
There are several mathematical definitions of skewness. One common expression to calculate the skewness of the dataset 𝑥 with 𝑛 elements is (𝑛² / ((𝑛 − 1)(𝑛 − 2))) (Σᵢ(𝑥ᵢ − mean(𝑥))³ / (𝑛𝑠³)). A simpler expression is Σᵢ(𝑥ᵢ − mean(𝑥))³ 𝑛 / ((𝑛 − 1)(𝑛 − 2)𝑠³), where 𝑖 = 1, 2, …, 𝑛 and mean(𝑥) is the sample mean of 𝑥. The skewness defined like this is called the adjusted Fisher-Pearson standardized moment coefficient.
The previous figure showed two datasets that were quite symmetrical. In other words, their points had similar distances from the mean. In contrast, the following image illustrates two asymmetrical sets:
The first set is represented by the green dots and the second with the white ones. Usually, negative skewness values indicate that there’s a dominant tail on the left side, which you can see with the first set. Positive skewness values correspond to a longer or fatter tail on the right side, which you can see in the second set. If the skewness is close to 0 (for example, between −0.5 and 0.5), then the dataset is considered quite symmetrical.
Once you’ve calculated the size of your dataset n, the sample mean mean_, and the standard deviation std_, you can get the sample skewness with pure Python:
>>> x=[8.0,1,2.5,4,28.0]>>> n=len(x)>>> mean_=sum(x)/n>>> var_=sum((item-mean_)**2foriteminx)/(n-1)>>> std_=var_**0.5>>> skew_=(sum((item-mean_)**3foriteminx)... *n/((n-1)*(n-2)*std_**3))>>> skew_1.9470432273905929The skewness is positive, so x has a right-side tail.
You can also calculate the sample skewness with scipy.stats.skew():
>>> y,y_with_nan=np.array(x),np.array(x_with_nan)>>> scipy.stats.skew(y,bias=False)1.9470432273905927>>> scipy.stats.skew(y_with_nan,bias=False)nanThe obtained result is the same as the pure Python implementation. The parameter bias is set to False to enable the corrections for statistical bias. The optional parameter nan_policy can take the values 'propagate', 'raise', or 'omit'. It allows you to control how you’ll handle nan values.
Pandas Series objects have the method .skew() that also returns the skewness of a dataset:
>>> z,z_with_nan=pd.Series(x),pd.Series(x_with_nan)>>> z.skew()1.9470432273905924>>> z_with_nan.skew()1.9470432273905924Like other methods, .skew() ignores nan values by default, because of the default value of the optional parameter skipna.
Percentiles
The sample 𝑝 percentile is the element in the dataset such that 𝑝% of the elements in the dataset are less than or equal to that value. Also, (100 − 𝑝)% of the elements are greater than or equal to that value. If there are two such elements in the dataset, then the sample 𝑝 percentile is their arithmetic mean. Each dataset has three quartiles, which are the percentiles that divide the dataset into four parts:
- The first quartile is the sample 25th percentile. It divides roughly 25% of the smallest items from the rest of the dataset.
- The second quartile is the sample 50th percentile or the median. Approximately 25% of the items lie between the first and second quartiles and another 25% between the second and third quartiles.
- The third quartile is the sample 75th percentile. It divides roughly 25% of the largest items from the rest of the dataset.
Each part has approximately the same number of items. If you want to divide your data into several intervals, then you can use statistics.quantiles():
>>> x=[-5.0,-1.1,0.1,2.0,8.0,12.8,21.0,25.8,41.0]>>> statistics.quantiles(x,n=2)[8.0]>>> statistics.quantiles(x,n=4,method='inclusive')[0.1, 8.0, 21.0]In this example, 8.0 is the median of x, while 0.1 and 21.0 are the sample 25th and 75th percentiles, respectively. The parameter n defines the number of resulting equal-probability percentiles, and method determines how to calculate them.
Note:statistics.quantiles() is introduced in Python 3.8.
You can also use np.percentile() to determine any sample percentile in your dataset. For example, this is how you can find the 5th and 95th percentiles:
>>> y=np.array(x)>>> np.percentile(y,5)-3.44>>> np.percentile(y,95)34.919999999999995percentile() takes several arguments. You have to provide the dataset as the first argument and the percentile value as the second. The dataset can be in the form of a NumPy array, list, tuple, or similar data structure. The percentile can be a number between 0 and 100 like in the example above, but it can also be a sequence of numbers:
>>> np.percentile(y,[25,50,75])array([ 0.1, 8. , 21. ])>>> np.median(y)8.0This code calculates the 25th, 50th, and 75th percentiles all at once. If the percentile value is a sequence, then percentile() returns a NumPy array with the results. The first statement returns the array of quartiles. The second statement returns the median, so you can confirm it’s equal to the 50th percentile, which is 8.0.
If you want to ignore nan values, then use np.nanpercentile() instead:
>>> y_with_nan=np.insert(y,2,np.nan)>>> y_with_nanarray([-5. , -1.1, nan, 0.1, 2. , 8. , 12.8, 21. , 25.8, 41. ])>>> np.nanpercentile(y_with_nan,[25,50,75])array([ 0.1, 8. , 21. ])That’s how you can avoid nan values.
NumPy also offers you very similar functionality in quantile() and nanquantile(). If you use them, then you’ll need to provide the quantile values as the numbers between 0 and 1 instead of percentiles:
>>> np.quantile(y,0.05)-3.44>>> np.quantile(y,0.95)34.919999999999995>>> np.quantile(y,[0.25,0.5,0.75])array([ 0.1, 8. , 21. ])>>> np.nanquantile(y_with_nan,[0.25,0.5,0.75])array([ 0.1, 8. , 21. ])The results are the same as in the previous examples, but here your arguments are between 0 and 1. In other words, you passed 0.05 instead of 5 and 0.95 instead of 95.
pd.Series objects have the method .quantile():
>>> z,z_with_nan=pd.Series(y),pd.Series(y_with_nan)>>> z.quantile(0.05)-3.44>>> z.quantile(0.95)34.919999999999995>>> z.quantile([0.25,0.5,0.75])0.25 0.10.50 8.00.75 21.0dtype: float64>>> z_with_nan.quantile([0.25,0.5,0.75])0.25 0.10.50 8.00.75 21.0dtype: float64.quantile() also needs you to provide the quantile value as the argument. This value can be a number between 0 and 1 or a sequence of numbers. In the first case, .quantile() returns a scalar. In the second case, it returns a new Series holding the results.
Ranges
The range of data is the difference between the maximum and minimum element in the dataset. You can get it with the function np.ptp():
>>> np.ptp(y)46.0>>> np.ptp(z)46.0>>> np.ptp(y_with_nan)nan>>> np.ptp(z_with_nan)46.0This function returns nan if there are nan values in your NumPy array. If you use a Pandas Series object, then it will return a number.
Alternatively, you can use built-in Python, NumPy, or Pandas functions and methods to calculate the maxima and minima of sequences:
max()andmin()from the Python standard libraryamax()andamin()from NumPynanmax()andnanmin()from NumPy to ignorenanvalues.max()and.min()from NumPy.max()and.min()from Pandas to ignorenanvalues by default
Here are some examples of how you would use these routines:
>>> np.amax(y)-np.amin(y)46.0>>> np.nanmax(y_with_nan)-np.nanmin(y_with_nan)46.0>>> y.max()-y.min()46.0>>> z.max()-z.min()46.0>>> z_with_nan.max()-z_with_nan.min()46.0That’s how you get the range of data.
The interquartile range is the difference between the first and third quartile. Once you calculate the quartiles, you can take their difference:
>>> quartiles=np.quantile(y,[0.25,0.75])>>> quartiles[1]-quartiles[0]20.9>>> quartiles=z.quantile([0.25,0.75])>>> quartiles[0.75]-quartiles[0.25]20.9Note that you access the values in a Pandas Series object with the labels 0.75 and 0.25.
Summary of Descriptive Statistics
SciPy and Pandas offer useful routines to quickly get descriptive statistics with a single function or method call. You can use scipy.stats.describe() like this:
>>> result=scipy.stats.describe(y,ddof=1,bias=False)>>> resultDescribeResult(nobs=9, minmax=(-5.0, 41.0), mean=11.622222222222222, variance=228.75194444444446, skewness=0.9249043136685094, kurtosis=0.14770623629658886)You have to provide the dataset as the first argument. The argument can be a NumPy array, list, tuple, or similar data structure. You can omit ddof=1 since it’s the default and only matters when you’re calculating the variance. You can pass bias=False to force correcting the skewness and kurtosis for statistical bias.
Note: The optional parameter nan_policy can take the values 'propagate' (default), 'raise' (an error), or 'omit'. This parameter allows you to control what’s happening when there are nan values.
describe() returns an object that holds the following descriptive statistics:
nobs: the number of observations or elements in your datasetminmax: the tuple with the minimum and maximum values of your datasetmean: the mean of your datasetvariance: the variance of your datasetskewness: the skewness of your datasetkurtosis: the kurtosis of your dataset
You can access particular values with dot notation:
>>> result.nobs9>>> result.minmax[0]# Min-5.0>>> result.minmax[1]# Max41.0>>> result.mean11.622222222222222>>> result.variance228.75194444444446>>> result.skewness0.9249043136685094>>> result.kurtosis0.14770623629658886With SciPy, you’re just one function call away from a descriptive statistics summary for your dataset.
Pandas has similar, if not better, functionality. Series objects have the method .describe():
>>> result=z.describe()>>> resultcount 9.000000mean 11.622222std 15.124548min -5.00000025% 0.10000050% 8.00000075% 21.000000max 41.000000dtype: float64It returns a new Series that holds the following:
count: the number of elements in your datasetmean: the mean of your datasetstd: the standard deviation of your datasetminandmax: the minimum and maximum values of your dataset25%,50%, and75%: the quartiles of your dataset
If you want the resulting Series object to contain other percentiles, then you should specify the value of the optional parameter percentiles. You can access each item of result with its label:
>>> result['mean']11.622222222222222>>> result['std']15.12454774346805>>> result['min']-5.0>>> result['max']41.0>>> result['25%']0.1>>> result['50%']8.0>>> result['75%']21.0That’s how you can get descriptive statistics of a Series object with a single method call using Pandas.
Measures of Correlation Between Pairs of Data
You’ll often need to examine the relationship between the corresponding elements of two variables in a dataset. Say there are two variables, 𝑥 and 𝑦, with an equal number of elements, 𝑛. Let 𝑥₁ from 𝑥 correspond to 𝑦₁ from 𝑦, 𝑥₂ from 𝑥 to 𝑦₂ from 𝑦, and so on. You can then say that there are 𝑛 pairs of corresponding elements: (𝑥₁, 𝑦₁), (𝑥₂, 𝑦₂), and so on.
You’ll see the following measures of correlation between pairs of data:
- Positive correlation exists when larger values of 𝑥 correspond to larger values of 𝑦 and vice versa.
- Negative correlation exists when larger values of 𝑥 correspond to smaller values of 𝑦 and vice versa.
- Weak or no correlation exists if there is no such apparent relationship.
The following figure shows examples of negative, weak, and positive correlation:
The plot on the left with the red dots shows negative correlation. The plot in the middle with the green dots shows weak correlation. Finally, the plot on the right with the blue dots shows positive correlation.
Note: There’s one important thing you should always have in mind when working with correlation among a pair of variables, and that’s that correlation is not a measure or indicator of causation, but only of association!
The two statistics that measure the correlation between datasets are covariance and the correlation coefficient. Let’s define some data to work with these measures. You’ll create two Python lists and use them to get corresponding NumPy arrays and Pandas Series:
>>> x=list(range(-10,11))>>> y=[0,2,2,2,2,3,3,6,7,4,7,6,6,9,4,5,5,10,11,12,14]>>> x_,y_=np.array(x),np.array(y)>>> x__,y__=pd.Series(x_),pd.Series(y_)Now that you have the two variables, you can start exploring the relationship between them.
Covariance
The sample covariance is a measure that quantifies the strength and direction of a relationship between a pair of variables:
- If the correlation is positive, then the covariance is positive, as well. A stronger relationship corresponds to a higher value of the covariance.
- If the correlation is negative, then the covariance is negative, as well. A stronger relationship corresponds to a lower (or higher absolute) value of the covariance.
- If the correlation is weak, then the covariance is close to zero.
The covariance of the variables 𝑥 and 𝑦 is mathematically defined as 𝑠ˣʸ = Σᵢ (𝑥ᵢ − mean(𝑥)) (𝑦ᵢ − mean(𝑦)) / (𝑛 − 1), where 𝑖 = 1, 2, …, 𝑛, mean(𝑥) is the sample mean of 𝑥, and mean(𝑦) is the sample mean of 𝑦. It follows that the covariance of two identical variables is actually the variance: 𝑠ˣˣ = Σᵢ(𝑥ᵢ − mean(𝑥))² / (𝑛 − 1) = (𝑠ˣ)² and 𝑠ʸʸ = Σᵢ(𝑦ᵢ − mean(𝑦))² / (𝑛 − 1) = (𝑠ʸ)².
This is how you can calculate the covariance in pure Python:
>>> n=len(x)>>> mean_x,mean_y=sum(x)/n,sum(y)/n>>> cov_xy=(sum((x[k]-mean_x)*(y[k]-mean_y)forkinrange(n))... /(n-1))>>> cov_xy19.95First, you have to find the mean of x and y. Then, you apply the mathematical formula for the covariance.
NumPy has the function cov() that returns the covariance matrix:
>>> cov_matrix=np.cov(x_,y_)>>> cov_matrixarray([[38.5 , 19.95 ], [19.95 , 13.91428571]])Note that cov() has the optional parameters bias, which defaults to False, and ddof, which defaults to None. Their default values are suitable for getting the sample covariance matrix. The upper-left element of the covariance matrix is the covariance of x and x, or the variance of x. Similarly, the lower-right element is the covariance of y and y, or the variance of y. You can check to see that this is true:
>>> x_.var(ddof=1)38.5>>> y_.var(ddof=1)13.914285714285711As you can see, the variances of x and y are equal to cov_matrix[0, 0] and cov_matrix[1, 1], respectively.
The other two elements of the covariance matrix are equal and represent the actual covariance between x and y:
>>> cov_xy=cov_matrix[0,1]>>> cov_xy19.95>>> cov_xy=cov_matrix[1,0]>>> cov_xy19.95You’ve obtained the same value of the covariance with np.cov() as with pure Python.
Pandas Series have the method .cov() that you can use to calculate the covariance:
>>> cov_xy=x__.cov(y__)>>> cov_xy19.95>>> cov_xy=y__.cov(x__)>>> cov_xy19.95Here, you call .cov() on one Series object and pass the other object as the first argument.
Correlation Coefficient
The correlation coefficient, or Pearson product-moment correlation coefficient, is denoted by the symbol 𝑟. The coefficient is another measure of the correlation between data. You can think of it as a standardized covariance. Here are some important facts about it:
- The value 𝑟 > 0 indicates positive correlation.
- The value 𝑟 < 0 indicates negative correlation.
- The value r = 1 is the maximum possible value of 𝑟. It corresponds to a perfect positive linear relationship between variables.
- The value r = −1 is the minimum possible value of 𝑟. It corresponds to a perfect negative linear relationship between variables.
- The value r ≈ 0, or when 𝑟 is around zero, means that the correlation between variables is weak.
The mathematical formula for the correlation coefficient is 𝑟 = 𝑠ˣʸ / (𝑠ˣ𝑠ʸ) where 𝑠ˣ and 𝑠ʸ are the standard deviations of 𝑥 and 𝑦 respectively. If you have the means (mean_x and mean_y) and standard deviations (std_x, std_y) for the datasets x and y, as well as their covariance cov_xy, then you can calculate the correlation coefficient with pure Python:
>>> var_x=sum((item-mean_x)**2foriteminx)/(n-1)>>> var_y=sum((item-mean_y)**2foriteminy)/(n-1)>>> std_x,std_y=var_x**0.5,var_y**0.5>>> r=cov_xy/(std_x*std_y)>>> r0.861950005631606You’ve got the variable r that represents the correlation coefficient.
scipy.stats has the routine pearsonr() that calculates the correlation coefficient and the 𝑝-value:
>>> r,p=scipy.stats.pearsonr(x_,y_)>>> r0.861950005631606>>> p5.122760847201171e-07pearsonr() returns a tuple with two numbers. The first one is 𝑟 and the second is the 𝑝-value.
Similar to the case of the covariance matrix, you can apply np.corrcoef() with x_ and y_ as the arguments and get the correlation coefficient matrix:
>>> corr_matrix=np.corrcoef(x_,y_)>>> corr_matrixarray([[1. , 0.86195001], [0.86195001, 1. ]])The upper-left element is the correlation coefficient between x_ and x_. The lower-right element is the correlation coefficient between y_ and y_. Their values are equal to 1.0. The other two elements are equal and represent the actual correlation coefficient between x_ and y_:
>>> r=corr_matrix[0,1]>>> r0.8619500056316061>>> r=corr_matrix[1,0]>>> r0.861950005631606Of course, the result is the same as with pure Python and pearsonr().
You can get the correlation coefficient with scipy.stats.linregress():
>>> scipy.stats.linregress(x_,y_)LinregressResult(slope=0.5181818181818181, intercept=5.714285714285714, rvalue=0.861950005631606, pvalue=5.122760847201164e-07, stderr=0.06992387660074979)linregress() takes x_ and y_, performs linear regression, and returns the results. slope and intercept define the equation of the regression line, while rvalue is the correlation coefficient. To access particular values from the result of linregress(), including the correlation coefficient, use dot notation:
>>> result=scipy.stats.linregress(x_,y_)>>> r=result.rvalue>>> r0.861950005631606That’s how you can perform linear regression and obtain the correlation coefficient.
Pandas Series have the method .corr() for calculating the correlation coefficient:
>>> r=x__.corr(y__)>>> r0.8619500056316061>>> r=y__.corr(x__)>>> r0.861950005631606You should call .corr() on one Series object and pass the other object as the first argument.
Working With 2D Data
Statisticians often work with 2D data. Here are some examples of 2D data formats:
- Database tables
- CSV files
- Excel, Calc, and Google spreadsheets
NumPy and SciPy provide a comprehensive means to work with 2D data. Pandas has the class DataFrame specifically to handle 2D labeled data.
Axes
Start by creating a 2D NumPy array:
>>> a=np.array([[1,1,1],... [2,3,1],... [4,9,2],... [8,27,4],... [16,1,1]])>>> aarray([[ 1, 1, 1], [ 2, 3, 1], [ 4, 9, 2], [ 8, 27, 4], [16, 1, 1]])Now you have a 2D dataset, which you’ll use in this section. You can apply Python statistics functions and methods to it just as you would to 1D data:
>>> np.mean(a)5.4>>> a.mean()5.4>>> np.median(a)2.0>>> a.var(ddof=1)53.40000000000001As you can see, you get statistics (like the mean, median, or variance) across all data in the array a. Sometimes, this behavior is what you want, but in some cases, you’ll want these quantities calculated for each row or column of your 2D array.
The functions and methods you’ve used so far have one optional parameter called axis, which is essential for handling 2D data. axis can take on any of the following values:
axis=Nonesays to calculate the statistics across all data in the array. The examples above work like this. This behavior is often the default in NumPy.axis=0says to calculate the statistics across all rows, that is, for each column of the array. This behavior is often the default for SciPy statistical functions.axis=1says to calculate the statistics across all columns, that is, for each row of the array.
Let’s see axis=0 in action with np.mean():
>>> np.mean(a,axis=0)array([6.2, 8.2, 1.8])>>> a.mean(axis=0)array([6.2, 8.2, 1.8])The two statements above return new NumPy arrays with the mean for each column of a. In this example, the mean of the first column is 6.2. The second column has the mean 8.2, while the third has 1.8.
If you provide axis=1 to mean(), then you’ll get the results for each row:
>>> np.mean(a,axis=1)array([ 1., 2., 5., 13., 6.])>>> a.mean(axis=1)array([ 1., 2., 5., 13., 6.])As you can see, the first row of a has the mean 1.0, the second 2.0, and so on.
Note: You can extend these rules to multi-dimensional arrays, but that’s beyond the scope of this tutorial. Feel free to dive into this topic on your own!
The parameter axis works the same way with other NumPy functions and methods:
>>> np.median(a,axis=0)array([4., 3., 1.])>>> np.median(a,axis=1)array([1., 2., 4., 8., 1.])>>> a.var(axis=0,ddof=1)array([ 37.2, 121.2, 1.7])>>> a.var(axis=1,ddof=1)array([ 0., 1., 13., 151., 75.])You’ve got the medians and sample variations for all columns (axis=0) and rows (axis=1) of the array a.
This is very similar when you work with SciPy statistics functions. But remember that in this case, the default value for axis is 0:
>>> scipy.stats.gmean(a)# Default: axis=0array([4. , 3.73719282, 1.51571657])>>> scipy.stats.gmean(a,axis=0)array([4. , 3.73719282, 1.51571657])If you omit axis or provide axis=0, then you’ll get the result across all rows, that is, for each column. For example, the first column of a has a geometric mean of 4.0, and so on.
If you specify axis=1, then you’ll get the calculations across all columns, that is for each row:
>>> scipy.stats.gmean(a,axis=1)array([1. , 1.81712059, 4.16016765, 9.52440631, 2.5198421 ])In this example, the geometric mean of the first row of a is 1.0. For the second row, it’s approximately 1.82, and so on.
If you want statistics for the entire dataset, then you have to provide axis=None:
>>> scipy.stats.gmean(a,axis=None)2.829705017016332The geometric mean of all the items in the array a is approximately 2.83.
You can get a Python statistics summary with a single function call for 2D data with scipy.stats.describe(). It works similar to 1D arrays, but you have to be careful with the parameter axis:
>>> scipy.stats.describe(a,axis=None,ddof=1,bias=False)DescribeResult(nobs=15, minmax=(1, 27), mean=5.4, variance=53.40000000000001, skewness=2.264965290423389, kurtosis=5.212690982795767)>>> scipy.stats.describe(a,ddof=1,bias=False)# Default: axis=0DescribeResult(nobs=5, minmax=(array([1, 1, 1]), array([16, 27, 4])), mean=array([6.2, 8.2, 1.8]), variance=array([ 37.2, 121.2, 1.7]), skewness=array([1.32531471, 1.79809454, 1.71439233]), kurtosis=array([1.30376344, 3.14969121, 2.66435986]))>>> scipy.stats.describe(a,axis=1,ddof=1,bias=False)DescribeResult(nobs=3, minmax=(array([1, 1, 2, 4, 1]), array([ 1, 3, 9, 27, 16])), mean=array([ 1., 2., 5., 13., 6.]), variance=array([ 0., 1., 13., 151., 75.]), skewness=array([0. , 0. , 1.15206964, 1.52787436, 1.73205081]), kurtosis=array([-3. , -1.5, -1.5, -1.5, -1.5]))When you provide axis=None, you get the summary across all data. Most results are scalars. If you set axis=0 or omit it, then the return value is the summary for each column. So, most results are the arrays with the same number of items as the number of columns. If you set axis=1, then describe() returns the summary for all rows.
You can get a particular value from the summary with dot notation:
>>> result=scipy.stats.describe(a,axis=1,ddof=1,bias=False)>>> result.meanarray([ 1., 2., 5., 13., 6.])That’s how you can see a statistics summary for a 2D array with a single function call.
DataFrames
The class DataFrame is one of the fundamental Pandas data types. It’s very comfortable to work with because it has labels for rows and columns. Use the array a and create a DataFrame:
>>> row_names=['first','second','third','fourth','fifth']>>> col_names=['A','B','C']>>> df=pd.DataFrame(a,index=row_names,columns=col_names)>>> df A B Cfirst 1 1 1second 2 3 1third 4 9 2fourth 8 27 4fifth 16 1 1In practice, the names of the columns matter and should be descriptive. The names of the rows are sometimes specified automatically as 0, 1, and so on. You can specify them explicitly with the parameter index, though you’re free to omit index if you like.
DataFrame methods are very similar to Series methods, though the behavior is different. If you call Python statistics methods without arguments, then the DataFrame will return the results for each column:
>>> df.mean()A 6.2B 8.2C 1.8dtype: float64>>> df.var()A 37.2B 121.2C 1.7dtype: float64What you get is a new Series that holds the results. In this case, the Series holds the mean and variance for each column. If you want the results for each row, then just specify the parameter axis=1:
>>> df.mean(axis=1)first 1.0second 2.0third 5.0fourth 13.0fifth 6.0dtype: float64>>> df.var(axis=1)first 0.0second 1.0third 13.0fourth 151.0fifth 75.0dtype: float64The result is a Series with the desired quantity for each row. The labels 'first', 'second', and so on refer to the different rows.
You can isolate each column of a DataFrame like this:
>>> df['A']first 1second 2third 4fourth 8fifth 16Name: A, dtype: int64Now, you have the column 'A' in the form of a Series object and you can apply the appropriate methods:
>>> df['A'].mean()6.2>>> df['A'].var()37.20000000000001That’s how you can obtain the statistics for a single column.
Sometimes, you might want to use a DataFrame as a NumPy array and apply some function to it. It’s possible to get all data from a DataFrame with .values or .to_numpy():
>>> df.valuesarray([[ 1, 1, 1], [ 2, 3, 1], [ 4, 9, 2], [ 8, 27, 4], [16, 1, 1]])>>> df.to_numpy()array([[ 1, 1, 1], [ 2, 3, 1], [ 4, 9, 2], [ 8, 27, 4], [16, 1, 1]])df.values and df.to_numpy() give you a NumPy array with all items from the DataFrame without row and column labels. Note that df.to_numpy() is more flexible because you can specify the data type of items and whether you want to use the existing data or copy it.
Like Series, DataFrame objects have the method .describe() that returns another DataFrame with the statistics summary for all columns:
>>> df.describe() A B Ccount 5.00000 5.000000 5.00000mean 6.20000 8.200000 1.80000std 6.09918 11.009087 1.30384min 1.00000 1.000000 1.0000025% 2.00000 1.000000 1.0000050% 4.00000 3.000000 1.0000075% 8.00000 9.000000 2.00000max 16.00000 27.000000 4.00000The summary contains the following results:
count: the number of items in each columnmean: the mean of each columnstd: the standard deviationminandmax: the minimum and maximum values25%,50%, and75%: the percentiles
If you want the resulting DataFrame object to contain other percentiles, then you should specify the value of the optional parameter percentiles.
You can access each item of the summary like this:
>>> df.describe().at['mean','A']6.2>>> df.describe().at['50%','B']3.0That’s how you can get descriptive Python statistics in one Series object with a single Pandas method call.
Visualizing Data
In addition to calculating the numerical quantities like mean, median, or variance, you can use visual methods to present, describe, and summarize data. In this section, you’ll learn how to present your data visually using the following graphs:
- Box plots
- Histograms
- Pie charts
- Bar charts
- X-Y plots
- Heatmaps
matplotlib.pyplot is a very convenient and widely-used library, though it’s not the only Python library available for this purpose. You can import it like this:
>>> importmatplotlib.pyplotasplt>>> plt.style.use('ggplot')Now, you have matplotlib.pyplot imported and ready for use. The second statement sets the style for your plots by choosing colors, line widths, and other stylistic elements. You’re free to omit these if you’re satisfied with the default style settings.
Note: This section focuses on representing data and keeps stylistic settings to a minimum. You’ll see links to the official documentation for used routines from matplotlib.pyplot, so you can explore the options that you won’t see here.
You’ll use pseudo-random numbers to get data to work with. You don’t need knowledge on random numbers to be able to understand this section. You just need some arbitrary numbers, and pseudo-random generators are a convenient tool to get them. The module np.random generates arrays of pseudo-random numbers:
- Normally distributed numbers are generated with
np.random.randn(). - Uniformly distributed integers are generated with
np.random.randint().
NumPy 1.17 introduced another module for pseudo-random number generation. To learn more about it, check the official documentation.
Box Plots
The box plot is an excellent tool to visually represent descriptive statistics of a given dataset. It can show the range, interquartile range, median, mode, outliers, and all quartiles. First, create some data to represent with a box plot:
>>> np.random.seed(seed=0)>>> x=np.random.randn(1000)>>> y=np.random.randn(100)>>> z=np.random.randn(10)The first statement sets the seed of the NumPy random number generator with seed(), so you can get the same results each time you run the code. You don’t have to set the seed, but if you don’t specify this value, then you’ll get different results each time.
The other statements generate three NumPy arrays with normally distributed pseudo-random numbers. x refers to the array with 1000 items, y has 100, and z contains 10 items. Now that you have the data to work with, you can apply .boxplot() to get the box plot:
fig,ax=plt.subplots()ax.boxplot((x,y,z),vert=False,showmeans=True,meanline=True,labels=('x','y','z'),patch_artist=True,medianprops={'linewidth':2,'color':'purple'},meanprops={'linewidth':2,'color':'red'})plt.show()The parameters of .boxplot() define the following:
xis your data.vertsets the plot orientation to horizontal whenFalse. The default orientation is vertical.showmeansshows the mean of your data whenTrue.meanlinerepresents the mean as a line whenTrue. The default representation is a point.labels: the labels of your data.patch_artistdetermines how to draw the graph.medianpropsdenotes the properties of the line representing the median.meanpropsindicates the properties of the line or dot representing the mean.
There are other parameters, but their analysis is beyond the scope of this tutorial.
The code above produces an image like this:
You can see three box plots. Each of them corresponds to a single dataset (x, y, or z) and show the following:
- The mean is the red dashed line.
- The median is the purple line.
- The first quartile is the left edge of the blue rectangle.
- The third quartile is the right edge of the blue rectangle.
- The interquartile range is the length of the blue rectangle.
- The range contains everything from left to right.
- The outliers are the dots to the left and right.
A box plot can show so much information in a single figure!
Histograms
Histograms are particularly useful when there are a large number of unique values in a dataset. The histogram divides the values from a sorted dataset into intervals, also called bins. Often, all bins are of equal width, though this doesn’t have to be the case. The values of the lower and upper bounds of a bin are called the bin edges.
The frequency is a single value that corresponds to each bin. It’s the number of elements of the dataset with the values between the edges of the bin. By convention, all bins but the rightmost one are half-open. They include the values equal to the lower bounds, but exclude the values equal to the upper bounds. The rightmost bin is closed because it includes both bounds. If you divide a dataset with the bin edges 0, 5, 10, and 15, then there are three bins:
- The first and leftmost bin contains the values greater than or equal to 0 and less than 5.
- The second bin contains the values greater than or equal to 5 and less than 10.
- The third and rightmost bin contains the values greater than or equal to 10 and less than or equal to 15.
The function np.histogram() is a convenient way to get data for histograms:
>>> hist,bin_edges=np.histogram(x,bins=10)>>> histarray([ 9, 20, 70, 146, 217, 239, 160, 86, 38, 15])>>> bin_edgesarray([-3.04614305, -2.46559324, -1.88504342, -1.3044936 , -0.72394379, -0.14339397, 0.43715585, 1.01770566, 1.59825548, 2.1788053 , 2.75935511])It takes the array with your data and the number (or edges) of bins and returns two NumPy arrays:
histcontains the frequency or the number of items corresponding to each bin.bin_edgescontains the edges or bounds of the bin.
What histogram() calculates, .hist() can show graphically:
fig,ax=plt.subplots()ax.hist(x,bin_edges,cumulative=False)ax.set_xlabel('x')ax.set_ylabel('Frequency')plt.show()The first argument of .hist() is the sequence with your data. The second argument defines the edges of the bins. The third disables the option to create a histogram with cumulative values. The code above produces a figure like this:
You can see the bin edges on the horizontal axis and the frequencies on the vertical axis.
It’s possible to get the histogram with the cumulative numbers of items if you provide the argument cumulative=True to .hist():
fig,ax=plt.subplots()ax.hist(x,bin_edges,cumulative=True)ax.set_xlabel('x')ax.set_ylabel('Frequency')plt.show()This code yields the following figure:
It shows the histogram with the cumulative values. The frequency of the first and leftmost bin is the number of items in this bin. The frequency of the second bin is the sum of the numbers of items in the first and second bins. The other bins follow this same pattern. Finally, the frequency of the last and rightmost bin is the total number of items in the dataset (in this case, 1000). You can also directly draw a histogram with pd.Series.hist() using matplotlib in the background.
Pie Charts
Pie charts represent data with a small number of labels and given relative frequencies. They work well even with the labels that can’t be ordered (like nominal data). A pie chart is a circle divided into multiple slices. Each slice corresponds to a single distinct label from the dataset and has an area proportional to the relative frequency associated with that label.
Let’s define data associated to three labels:
>>> x,y,z=128,256,1024Now, create a pie chart with .pie():
fig,ax=plt.subplots()ax.pie((x,y,z),labels=('x','y','z'),autopct='%1.1f%%')plt.show()The first argument of .pie() is your data, and the second is the sequence of the corresponding labels. autopct defines the format of the relative frequencies shown on the figure. You’ll get a figure that looks like this:
The pie chart shows x as the smallest part of the circle, y as the next largest, and then z as the largest part. The percentages denote the relative size of each value compared to their sum.
Bar Charts
Bar charts also illustrate data that correspond to given labels or discrete numeric values. They can show the pairs of data from two datasets. Items of one set are the labels, while the corresponding items of the other are their frequencies. Optionally, they can show the errors related to the frequencies, as well.
The bar chart shows parallel rectangles called bars. Each bar corresponds to a single label and has a height proportional to the frequency or relative frequency of its label. Let’s generate three datasets, each with 21 items:
>>> x=np.arange(21)>>> y=np.random.randint(21,size=21)>>> err=np.random.randn(21)You use np.arange() to get x, or the array of consecutive integers from 0 to 20. You’ll use this to represent the labels. y is an array of uniformly distributed random integers, also between 0 and 20. This array will represent the frequencies. err contains normally distributed floating-point numbers, which are the errors. These values are optional.
You can create a bar chart with .bar() if you want vertical bars or .barh() if you’d like horizontal bars:
fig,ax=plt.subplots())ax.bar(x,y,yerr=err)ax.set_xlabel('x')ax.set_ylabel('y')plt.show()This code should produce the following figure:
The heights of the red bars correspond to the frequencies y, while the lengths of the black lines show the errors err. If you don’t want to include the errors, then omit the parameter yerr of .bar().
X-Y Plots
The x-y plot or scatter plot represents the pairs of data from two datasets. The horizontal x-axis shows the values from the set x, while the vertical y-axis shows the corresponding values from the set y. You can optionally include the regression line and the correlation coefficient. Let’s generate two datasets and perform linear regression with scipy.stats.linregress():
>>> x=np.arange(21)>>> y=5+2*x+2*np.random.randn(21)>>> slope,intercept,r,*__=scipy.stats.linregress(x,y)>>> line=f'Regression line: y={intercept:.2f}+{slope:.2f}x, r={r:.2f}'The dataset x is again the array with the integers from 0 to 20. y is calculated as a linear function of x distorted with some random noise.
linregress returns several values. You’ll need the slope and intercept of the regression line, as well as the correlation coefficient r. Then you can apply .plot() to get the x-y plot:
fig,ax=plt.subplots()ax.plot(x,y,linewidth=0,marker='s',label='Data points')ax.plot(x,intercept+slope*x,label=line)ax.set_xlabel('x')ax.set_ylabel('y')ax.legend(facecolor='white')plt.show()The result of the code above is this figure:
You can see the data points (x-y pairs) as red squares, as well as the blue regression line.
Heatmaps
A heatmap can be used to visually show a matrix. The colors represent the numbers or elements of the matrix. Heatmaps are particularly useful for illustrating the covariance and correlation matrices. You can create the heatmap for a covariance matrix with .imshow():
matrix=np.cov(x,y).round(decimals=2)fig,ax=plt.subplots()ax.imshow(matrix)ax.grid(False)ax.xaxis.set(ticks=(0,1),ticklabels=('x','y'))ax.yaxis.set(ticks=(0,1),ticklabels=('x','y'))ax.set_ylim(1.5,-0.5)foriinrange(2):forjinrange(2):ax.text(j,i,matrix[i,j],ha='center',va='center',color='w')plt.show()Here, the heatmap contains the labels 'x' and 'y' as well as the numbers from the covariance matrix. You’ll get a figure like this:
The yellow field represents the largest element from the matrix 130.34, while the purple one corresponds to the smallest element 38.5. The blue squares in between are associated with the value 69.9.
You can obtain the heatmap for the correlation coefficient matrix following the same logic:
matrix=np.corrcoef(x,y).round(decimals=2)fig,ax=plt.subplots()ax.imshow(matrix)ax.grid(False)ax.xaxis.set(ticks=(0,1),ticklabels=('x','y'))ax.yaxis.set(ticks=(0,1),ticklabels=('x','y'))ax.set_ylim(1.5,-0.5)foriinrange(2):forjinrange(2):ax.text(j,i,matrix[i,j],ha='center',va='center',color='w')plt.show()The result is the figure below:
The yellow color represents the value 1.0, and the purple color shows 0.99.
Conclusion
You now know the quantities that describe and summarize datasets and how to calculate them in Python. It’s possible to get descriptive statistics with pure Python code, but that’s rarely necessary. Usually, you’ll use some of the libraries created especially for this purpose:
- Use Python’s
statisticsfor the most important Python statistics functions. - Use NumPy to handle arrays efficiently.
- Use SciPy for additional Python statistics routines for NumPy arrays.
- Use Pandas to work with labeled datasets.
- Use Matplotlib to visualize data with plots, charts, and histograms.
In the era of big data and artificial intelligence, you must know how to calculate descriptive statistics measures. Now you’re ready to dive deeper into the world of data science and machine learning! If you have questions or comments, then please put them in the comments section below.
[ Improve Your Python With 🐍 Python Tricks 💌 – Get a short & sweet Python Trick delivered to your inbox every couple of days. >> Click here to learn more and see examples ]