My first Caktus project went live late in the summer of 2015. It's a community portal for users of an SMS-based product called RapidPro. The portal was built in the Wagtail CMS framework which has a lovely, intuitive admin interface and excellent documentation for developers and content editors. The code for our Wagtail-based project is all open sourced on GitHub.
For this community portal, we needed to allow users to create blog pages on our front-facing site without giving those same users any level of access to the actual CMS. We also didn't want outside users to have to learn a new CMS just to submit content.
We wanted a simple, one-stop form that guided users through entering their content and thanked them for submitting. After these outside users requested pages be published on the site, CMS content editors could then view, edit, and publish the pages through the Wagtail CMS.
Here's how we accomplished this in two steps. Karen Tracey and I both worked on this project and a lot of this code was guided by her Django wisdom.
Step 1: Use the RoutablePageMixin for our form page and thank you page
Now for a little background information on Wagtail. The Wagtail CMS framework allows you to create a model for each type of page on your site. For example, you might have one model for a blog page and another model for a blog index page that lists out your blog pages and allows you to search through blog pages. Each page model automatically connects to one template, based on a naming convention. For example, if your model is called BlogIndexPage, you would need to also have a template called blog_index_page.html, so that Wagtail knows how to find the related template. You don't have to write any views to use Wagtail out of the box.
However, in our case, we wanted users to submit a BlogPage entry which would be a child of a BlogIndexPage. Therefore, we wanted our BlogIndexPage model to route to itself, to a submission page, and to a "thank you" page.

This is where Wagtail's RoutablePageMixin came into play. Here's the relevant code from our BlogIndexPage model that routes the user from the list page to the submission page, then to the thank you page.
In models.py:
fromdjango.template.responseimportTemplateResponsefromwagtail.wagtailcore.modelsimportPagefromwagtail.wagtailcore.fieldsimportRichTextFieldfromwagtail.contrib.wagtailroutablepage.modelsimportRoutablePageMixin,routeclassBlogIndexPage(RoutablePageMixin,Page):intro=RichTextField(blank=True)submit_info=RichTextField(blank=True)thanks_info=RichTextField(blank=True)@route(r'^$')defbase(self,request):returnTemplateResponse(request,self.get_template(request),self.get_context(request))@route(r'^submit-blog/$')defsubmit(self,request):from.viewsimportsubmit_blogreturnsubmit_blog(request,self)@route(r'^submit-thank-you/$')defthanks(self,request):returnTemplateResponse(request,'portal_pages/thank_you.html',{"thanks_info":self.thanks_info})The base() method points us to the blog index page itself. Once we added the RoutablePageMixin, we had to explicitly define this method to pass the request, template, and context to the related template. If we weren't using this mixin, Wagtail would just route to the correct template based on the naming convention I described earlier.
The submit() method routes to our blog submission view. We decided to use the URL string "submit-blog/" but we could have called it anything. We have a view method submit_blog() defined in our views.py file that does the work of actually adding the page to the CMS.
The thanks() method routes to the thank you page (thank_you.html) and passes in content editable via the CMS in the variable thanks_info as defined in the BlogIndexPage model.
Step 2: Creating the form and view method to save the user-generated information
Here's the slightly trickier part, because we didn't find any documentation on adding pages to Wagtail programmatically. We found some of this code by digging deeper through the Wagtail repo and found the tests files especially helpful. Here are the relevant parts of our code.
In forms.py, we added a Django ModelForm.
classBlogForm(forms.ModelForm):classMeta:model=BlogPageIn views.py, we created a view method called submit_blog() that does a number of things.
- Imports the BlogForm form into the context of the page.
- Upon submission/post, saves the BlogForm with commit=False, so that it is not saved to the database, yet.
- Creates a slug based on the title the user entered with slugify(). This would normally be auto-generated and editable in the Wagtail CMS.
- Adds the unsaved BlogPage as a child to the BlogIndexPage (we passed in the reference to the index page in our routable submit() view method).
- Saves the page with the unpublish() command which both saves the uncommitted data to our CMS and marks it as a Draft for review.
- Saves the revision of the page so that we can later notify the Wagtail admins that a new page is waiting for their review with save_revision(submitted_for_moderation=True)
- Finally, this sends out email notifications to all the Wagtail admins with send_notification(blog.get_latest_revision().id, 'submitted', None). The None parameter in this function means do not exclude any Wagtail moderators.
defsubmit_blog(request,blog_index):form=BlogForm(data=request.POSTorNone,label_suffix='')ifrequest.method=='POST'andform.is_valid():blog_page=form.save(commit=False)blog_page.slug=slugify(blog_page.title)blog=blog_index.add_child(instance=blog_page)ifblog:blog.unpublish()# Submit page for moderation. This requires first saving a revision.blog.save_revision(submitted_for_moderation=True)# Then send the notification to all Wagtail moderators.send_notification(blog.get_latest_revision().id,'submitted',None)returnHttpResponseRedirect(blog_index.url+blog_index.reverse_subpage('thanks'))context={'form':form,'blog_index':blog_index,}returnrender(request,'portal_pages/blog_page_add.html',context)Final Thoughts and Some Screenshots

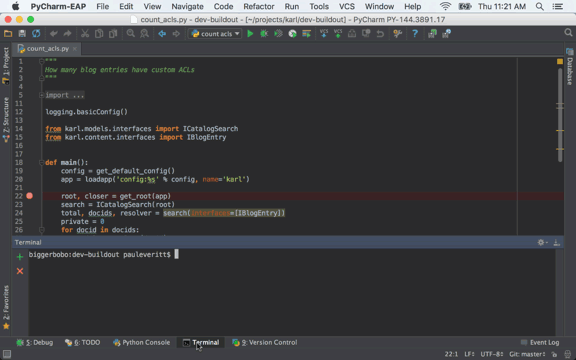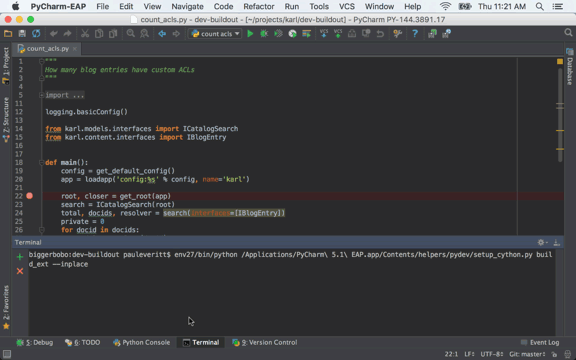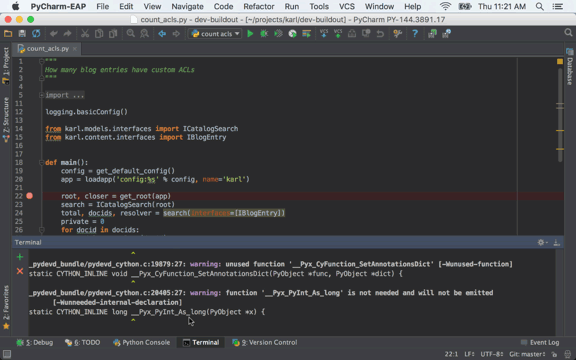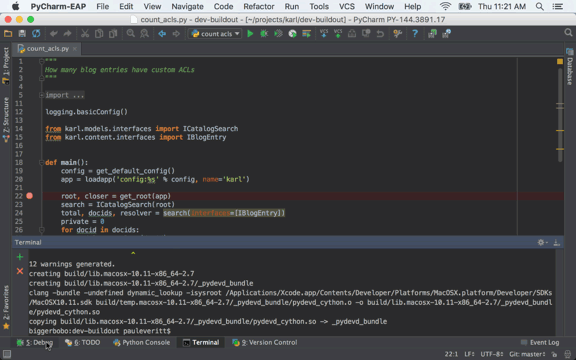
Front-end website for user submission of blog content.
Wagtail is very straightforward to use; we plan to use it on future projects. If you want to get started with Wagtail, the documentation is very thorough and well written. I also highly recommend downloading the open sourced demo site and getting that rolling in order to see how it's all hooked together.